1CXF
 
 | | COMPLEX OF A (D229N/E257Q) DOUBLE MUTANT CGTASE FROM BACILLUS CIRCULANS STRAIN 251 WITH MALTOTETRAOSE AT 120 K AND PH 9.1 OBTAINED AFTER SOAKING THE CRYSTAL WITH ALPHA-CYCLODEXTRIN | | Descriptor: | CALCIUM ION, CYCLODEXTRIN GLYCOSYLTRANSFERASE, Cyclohexakis-(1-4)-(alpha-D-glucopyranose), ... | | Authors: | Knegtel, R.M.A, Strokopytov, B.V, Dijkstra, B.W. | | Deposit date: | 1995-07-28 | | Release date: | 1995-12-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallographic studies of the interaction of cyclodextrin glycosyltransferase from Bacillus circulans strain 251 with natural substrates and products.
J.Biol.Chem., 270, 1995
|
|
1CXZ
 
 | | CRYSTAL STRUCTURE OF HUMAN RHOA COMPLEXED WITH THE EFFECTOR DOMAIN OF THE PROTEIN KINASE PKN/PRK1 | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, MAGNESIUM ION, PROTEIN (HIS-TAGGED TRANSFORMING PROTEIN RHOA(0-181)), ... | | Authors: | Maesaki, R, Ihara, K, Shimizu, T, Kuroda, S, Kaibuchi, K, Hakoshima, T. | | Deposit date: | 1999-08-31 | | Release date: | 1999-10-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structural basis of Rho effector recognition revealed by the crystal structure of human RhoA complexed with the effector domain of PKN/PRK1.
Mol.Cell, 4, 1999
|
|
6WC9
 
 | | Human open state TMEM175 in KCl | | Descriptor: | Endosomal/lysosomal potassium channel TMEM175, POTASSIUM ION | | Authors: | Oh, S, Paknejad, N, Hite, R.K. | | Deposit date: | 2020-03-30 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Gating and selectivity mechanisms for the lysosomal K + channel TMEM175.
Elife, 9, 2020
|
|
1CTZ
 
 | |
1CUO
 
 | | CRYSTAL STRUCTURE ANALYSIS OF ISOMER-2 AZURIN FROM METHYLOMONAS J | | Descriptor: | COPPER (II) ION, PROTEIN (AZURIN ISO-2) | | Authors: | Inoue, T, Nishio, N, Kai, Y, Suzuki, S, Kataoka, K. | | Deposit date: | 1999-08-21 | | Release date: | 2000-08-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The significance of the flexible loop in the azurin (Az-iso2) from the obligate methylotroph Methylomonas sp. strain J.
J.Mol.Biol., 333, 2003
|
|
1CY9
 
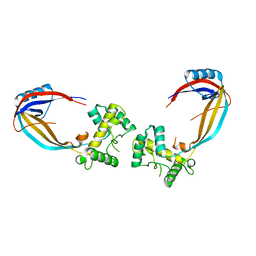 | |
1CZM
 
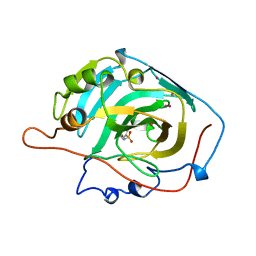 | |
1CVI
 
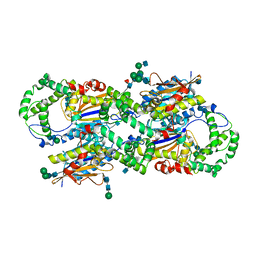 | | CRYSTAL STRUCTURE OF HUMAN PROSTATIC ACID PHOSPHATASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCINE, ... | | Authors: | Jakob, C.G, Lewinski, K, Kuciel, R, Ostrowski, W, Lebioda, L. | | Deposit date: | 1999-08-23 | | Release date: | 1999-08-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of human prostatic acid phosphatase .
Prostate, 42, 2000
|
|
1CWQ
 
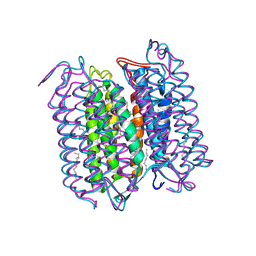 | | M INTERMEDIATE STRUCTURE OF THE WILD TYPE BACTERIORHODOPSIN IN COMBINATION WITH THE GROUND STATE STRUCTURE | | Descriptor: | BACTERIORHODOPSIN ("M" STATE INTERMEDIATE IN COMBINATION WITH GROUND STATE), HEXANE, N-OCTANE, ... | | Authors: | Sass, H.J, Berendzen, J, Neff, D, Gessenich, R, Ormos, P, Bueldt, G. | | Deposit date: | 1999-08-26 | | Release date: | 1999-10-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural alterations for proton translocation in the M state of wild-type bacteriorhodopsin.
Nature, 406, 2000
|
|
1CXI
 
 | |
1D01
 
 | | STRUCTURE OF TNF RECEPTOR ASSOCIATED FACTOR 2 IN COMPLEX WITH A HUMAN CD30 PEPTIDE | | Descriptor: | CD30 PEPTIDE, TUMOR NECROSIS FACTOR RECEPTOR ASSOCIATED FACTOR 2 | | Authors: | Ye, H, Park, Y.C, Kreishman, M, Kieff, E, Wu, H. | | Deposit date: | 1999-09-07 | | Release date: | 2003-12-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structural basis for the recognition of diverse receptor sequences by TRAF2.
Mol.Cell, 4, 1999
|
|
1D0U
 
 | |
1D1K
 
 | |
1D1U
 
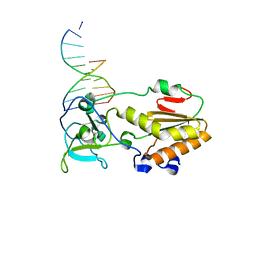 | | USE OF AN N-TERMINAL FRAGMENT FROM MOLONEY MURINE LEUKEMIA VIRUS REVERSE TRANSCRIPTASE TO FACILITATE CRYSTALLIZATION AND ANALYSIS OF A PSEUDO-16-MER DNA MOLECULE CONTAINING G-A MISPAIRS | | Descriptor: | DNA (5'-D(*AP*CP*GP*GP*CP*AP*CP*GP*AP*G)-3'), DNA (5'-D(*CP*TP*CP*GP*TP*G)-3'), PROTEIN (REVERSE TRANSCRIPTASE) | | Authors: | Cote, M.L, Yohannan, S, Georgiadis, M.M. | | Deposit date: | 1999-09-21 | | Release date: | 2000-04-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Use of an N-terminal fragment from moloney murine leukemia virus reverse transcriptase to facilitate crystallization and analysis of a pseudo-16-mer DNA molecule containing G-A mispairs.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1CY1
 
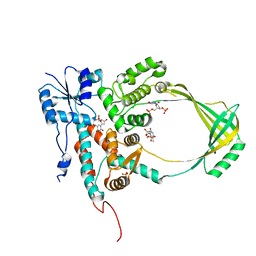 | | COMPLEX OF E.COLI DNA TOPOISOMERASE I WITH 5'PTPTPT | | Descriptor: | DNA TOPOISOMERASE I, PHOSPHATE ION, THYMIDINE-3',5'-DIPHOSPHATE, ... | | Authors: | Feinberg, H, Changela, A, Mondragon, A. | | Deposit date: | 1999-08-31 | | Release date: | 2000-03-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Protein-nucleotide interactions in E. coli DNA topoisomerase I.
Nat.Struct.Biol., 6, 1999
|
|
1D23
 
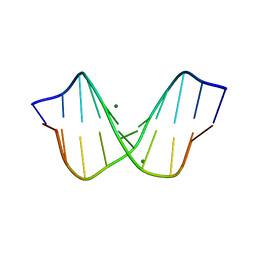 | | THE STRUCTURE OF B-HELICAL C-G-A-T-C-G-A-T-C-G AND COMPARISON WITH C-C-A-A-C-G-T-T-G-G. THE EFFECT OF BASE PAIR REVERSALS | | Descriptor: | DNA (5'-D(*CP*GP*AP*TP*CP*GP*AP*TP*CP*G)-3'), MAGNESIUM ION | | Authors: | Grzeskowiak, K, Yanagi, K, Prive, G.G, Dickerson, R.E. | | Deposit date: | 1991-05-29 | | Release date: | 1991-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structure of B-helical C-G-A-T-C-G-A-T-C-G and comparison with C-C-A-A-C-G-T-T-G-G. The effect of base pair reversals.
J.Biol.Chem., 266, 1991
|
|
1D29
 
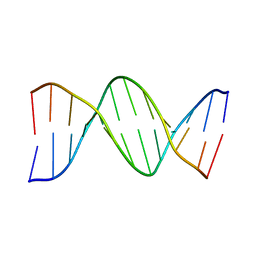 | |
1D2D
 
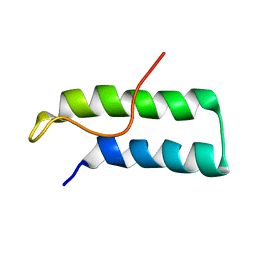 | | Hamster EprS second repeated element. NMR, 5 structures | | Descriptor: | TRNA SYNTHETASE | | Authors: | Cahuzac, B, Berthonneau, E, Birlirakis, N, Guittet, E, Mirande, M. | | Deposit date: | 1999-09-23 | | Release date: | 2000-05-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A recurrent RNA-binding domain is appended to eukaryotic aminoacyl-tRNA synthetases.
EMBO J., 19, 2000
|
|
1CEF
 
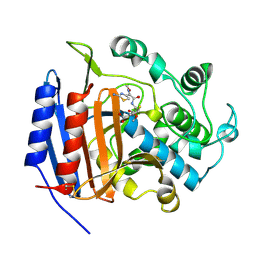 | | CEFOTAXIME COMPLEXED WITH THE STREPTOMYCES R61 DD-PEPTIDASE | | Descriptor: | CEFOTAXIME, C3' cleaved, open, ... | | Authors: | Knox, J.R, Kuzin, A.P. | | Deposit date: | 1995-01-12 | | Release date: | 1996-10-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Binding of cephalothin and cefotaxime to D-ala-D-ala-peptidase reveals a functional basis of a natural mutation in a low-affinity penicillin-binding protein and in extended-spectrum beta-lactamases.
Biochemistry, 34, 1995
|
|
1D36
 
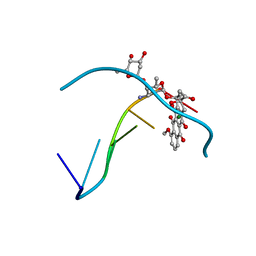 | | FACILE FORMATION OF A CROSSLINKED ADDUCT BETWEEN DNA AND THE DAUNORUBICIN DERIVATIVE MAR70 MEDIATED BY FORMALDEHYDE: MOLECULAR STRUCTURE OF THE MAR70-D(CGTNACG) COVALENT ADDUC | | Descriptor: | 4'-EPI-4'-(2-DEOXYFUCOSE)DAUNOMYCIN, DNA (5'-D(*CP*GP*TP*AP*CP*G)-3'), MAGNESIUM ION | | Authors: | Gao, Y.-G, Liaw, Y.-C, Li, Y.-K, Van Der Marel, G.A, Van Boom, J.H, Wang, A.H.-J. | | Deposit date: | 1991-04-23 | | Release date: | 1992-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Facile formation of a crosslinked adduct between DNA and the daunorubicin derivative MAR70 mediated by formaldehyde: molecular structure of the MAR70-d(CGTnACG) covalent adduct.
Proc.Natl.Acad.Sci.USA, 88, 1991
|
|
1D3W
 
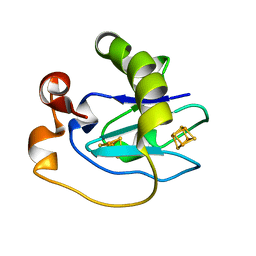 | | Crystal structure of ferredoxin 1 d15e mutant from azotobacter vinelandii at 1.7 angstrom resolution. | | Descriptor: | FE3-S4 CLUSTER, FERREDOXIN 1, IRON/SULFUR CLUSTER | | Authors: | Chen, K, Hirst, J, Camba, R, Bonagura, C.A, Stout, C.D, Burges, B.K, Armstrong, F.A. | | Deposit date: | 1999-10-01 | | Release date: | 1999-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Atomically defined mechanism for proton transfer to a buried redox centre in a protein.
Nature, 405, 2000
|
|
1D46
 
 | | DNA DODECAMER C-G-C-G-A-A-T-T-C-G-C-G/HOECHST 33258 COMPLEX:-100 DEGREES C, PIPERAZINE DOWN | | Descriptor: | 2'-(4-HYDROXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Quintana, J.R, Lipanov, A.A, Dickerson, R.E. | | Deposit date: | 1991-06-04 | | Release date: | 1992-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Low-temperature crystallographic analyses of the binding of Hoechst 33258 to the double-helical DNA dodecamer C-G-C-G-A-A-T-T-C-G-C-G.
Biochemistry, 30, 1991
|
|
1CZ7
 
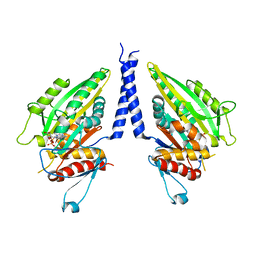 | | THE CRYSTAL STRUCTURE OF A MINUS-END DIRECTED MICROTUBULE MOTOR PROTEIN NCD REVEALS VARIABLE DIMER CONFORMATIONS | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, MICROTUBULE MOTOR PROTEIN NCD | | Authors: | Kozielski, F.K, De Bonis, S, Burmeister, W, Cohen-Addad, C, Wade, R. | | Deposit date: | 1999-09-01 | | Release date: | 1999-11-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The crystal structure of the minus-end-directed microtubule motor protein ncd reveals variable dimer conformations.
Structure Fold.Des., 7, 1999
|
|
1D00
 
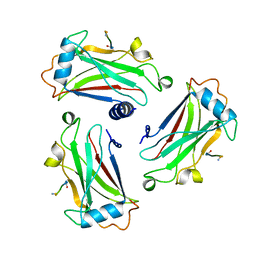 | | STRUCTURE OF TNF RECEPTOR ASSOCIATED FACTOR 2 IN COMPLEX WITH A 5-RESIDUE CD40 PEPTIDE | | Descriptor: | B-CELL SURFACE ANTIGEN CD40, TUMOR NECROSIS FACTOR RECEPTOR ASSOCIATED PROTEIN 2 | | Authors: | Ye, H, Park, Y.C, Kreishman, M, Kieff, E, Wu, H. | | Deposit date: | 1999-09-07 | | Release date: | 2000-03-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structural basis for the recognition of diverse receptor sequences by TRAF2.
Mol.Cell, 4, 1999
|
|
1D0W
 
 | |
