2NMY
 
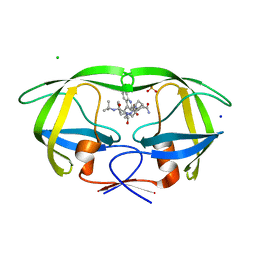 | | Crystal structure analysis of HIV-1 protease mutant V82A with a inhibitor saquinavir | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, CHLORIDE ION, PROTEASE, ... | | Authors: | Tie, Y, Kovalevsky, A.Y, Boross, P, Wang, Y.F, Ghosh, A.K, Tozser, J, Harrison, R.W, Weber, I.T. | | Deposit date: | 2006-10-23 | | Release date: | 2007-03-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic resolution crystal structures of HIV-1 protease and mutants V82A and I84V with saquinavir.
Proteins, 67, 2007
|
|
3MNC
 
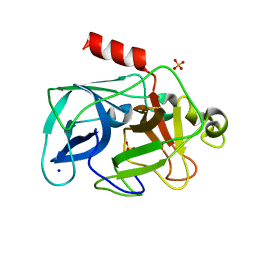 | | Investigation of global and local effects of radiation damage on porcine pancreatic elastase. Second stage of radiation damage | | Descriptor: | Chymotrypsin-like elastase family member 1, SODIUM ION, SULFATE ION | | Authors: | Petrova, T, Ginell, S, Kim, Y, Joachimiak, G, Joachimiak, A. | | Deposit date: | 2010-04-21 | | Release date: | 2010-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.119 Å) | | Cite: | X-ray-induced deterioration of disulfide bridges at atomic resolution.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
1B86
 
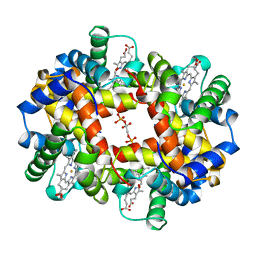 | | HUMAN DEOXYHAEMOGLOBIN-2,3-DIPHOSPHOGLYCERATE COMPLEX | | Descriptor: | (2R)-2,3-diphosphoglyceric acid, OXYGEN MOLECULE, PROTEIN (HEMOGLOBIN; ALPHA CHAIN), ... | | Authors: | Richard, V, Dodson, G.G, Mauguen, Y. | | Deposit date: | 1999-02-08 | | Release date: | 1999-02-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Human deoxyhaemoglobin-2,3-diphosphoglycerate complex low-salt structure at 2.5 A resolution.
J.Mol.Biol., 233, 1993
|
|
2NNQ
 
 | |
1B9S
 
 | | NOVEL AROMATIC INHIBITORS OF INFLUENZA VIRUS NEURAMINIDASE MAKE SELECTIVE INTERACTIONS WITH CONSERVED RESIDUES AND WATER MOLECULES IN THE ACTIVE SITE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(N-ACETYLAMINO)-3-[N-(2-ETHYLBUTANOYLAMINO)]BENZOIC ACID, CALCIUM ION, ... | | Authors: | Finley, J.B, Atigadda, V.R, Duarte, F, Zhao, J.J, Brouillette, W.J, Air, G.M, Luo, M. | | Deposit date: | 1999-02-15 | | Release date: | 1999-02-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Novel aromatic inhibitors of influenza virus neuraminidase make selective interactions with conserved residues and water molecules in the active site.
J.Mol.Biol., 293, 1999
|
|
1AZV
 
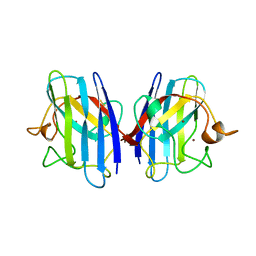 | | FAMILIAL ALS MUTANT G37R CUZNSOD (HUMAN) | | Descriptor: | COPPER (II) ION, COPPER/ZINC SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Hart, P.J, Liu, H, Pellegrini, M, Nersissian, A.M, Gralla, E.B, Valentine, J.S, Eisenberg, D. | | Deposit date: | 1997-11-21 | | Release date: | 1998-02-25 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Subunit asymmetry in the three-dimensional structure of a human CuZnSOD mutant found in familial amyotrophic lateral sclerosis.
Protein Sci., 7, 1998
|
|
1BB6
 
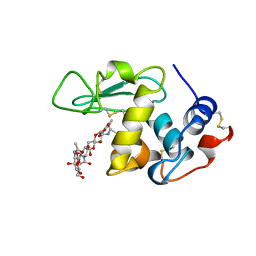 | |
2NAB
 
 | | Nizp1-C2HR zinc finger structure | | Descriptor: | ZINC ION, Zinc finger protein 496 | | Authors: | Berardi, A, Quilici, G, Spiliotopoulos, D, Corral-Rodriguez, M, Martin, F, Degano, M, Tonon, G, Musco, G. | | Deposit date: | 2015-12-22 | | Release date: | 2016-03-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for PHDVC5HCHNSD1-C2HRNizp1 interaction: implications for Sotos syndrome.
Nucleic Acids Res., 44, 2016
|
|
1BBZ
 
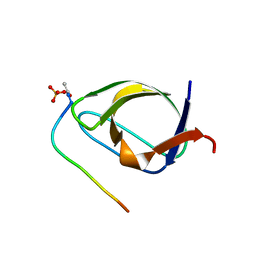 | |
2NA2
 
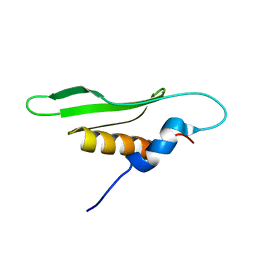 | |
1B2C
 
 | |
1BCJ
 
 | |
1B39
 
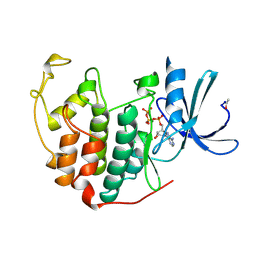 | | HUMAN CYCLIN-DEPENDENT KINASE 2 PHOSPHORYLATED ON THR 160 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PROTEIN (CELL DIVISION PROTEIN KINASE 2) | | Authors: | Brown, N.R, Noble, M.E.M, Lawrie, A.M, Morris, M.C, Tunnah, P, Divita, G, Johnson, L.N, Endicott, J.A. | | Deposit date: | 1998-12-17 | | Release date: | 1998-12-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Effects of phosphorylation of threonine 160 on cyclin-dependent kinase 2 structure and activity.
J.Biol.Chem., 274, 1999
|
|
1B4T
 
 | | H48C YEAST CU(II)/ZN SUPEROXIDE DISMUTASE ROOM TEMPERATURE (298K) STRUCTURE | | Descriptor: | CHLORIDE ION, COPPER (II) ION, PROTEIN (CU/ZN SUPEROXIDE DISMUTASE), ... | | Authors: | Hart, P.J, Balbirnie, M.M, Ogihara, N.L, Nersissian, A.M, Weiss, M.S, Valentine, J.S, Eisenberg, D. | | Deposit date: | 1998-12-23 | | Release date: | 1999-12-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A structure-based mechanism for copper-zinc superoxide dismutase.
Biochemistry, 38, 1999
|
|
3MLM
 
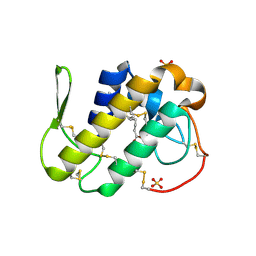 | | Crystal structure of Bn IV in complex with myristic acid: A Lys49 myotoxic phospholipase A2 from Bothrops neuwiedi venom | | Descriptor: | BN-IV Lys-49 Phospholipase A2, MYRISTIC ACID, SULFATE ION | | Authors: | Delatorre, P, Rocha, B.A.M, Cavada, B.S, Toyama, M.H, Toyama, D, Gadelha, C.A.A. | | Deposit date: | 2010-04-17 | | Release date: | 2011-05-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structure of Bn IV in complex with myristic acid: a Lys49 myotoxic phospholipase A2 from Bothrops neuwiedi venom.
Biochimie, 93, 2011
|
|
1BDA
 
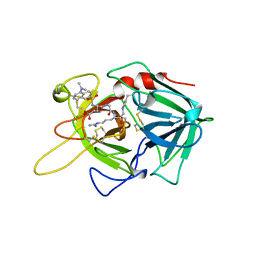 | |
7F68
 
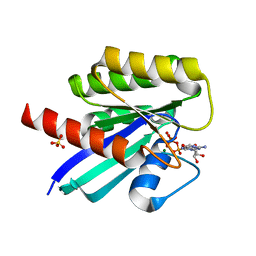 | | Crystal structure of N-ras S89D | | Descriptor: | GTPase NRas, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Li, Y, Sun, Q. | | Deposit date: | 2021-06-24 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Crystal structure of N-ras S89D
To Be Published
|
|
1BHZ
 
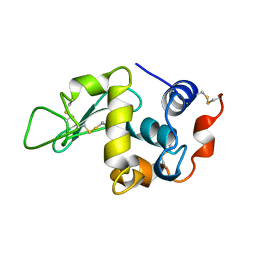 | | LOW TEMPERATURE MIDDLE RESOLUTION STRUCTURE OF HEN EGG WHITE LYSOZYME FROM MASC DATA | | Descriptor: | LYSOZYME | | Authors: | Ramin, M, Shepard, W, Fourme, R, Kahn, R. | | Deposit date: | 1998-06-10 | | Release date: | 1998-11-04 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Multiwavelength anomalous solvent contrast (MASC): derivation of envelope structure-factor amplitudes and comparison with model values.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
2NNB
 
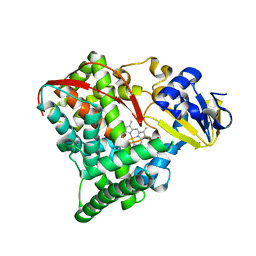 | | The Q403K mutant heme domain of flavocytochrome P450 BM3 | | Descriptor: | Bifunctional P-450:NADPH-P450 reductase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Clark, J.P, Anderson, J.L.R, Miles, C.S, Mowat, C.G, Reid, G.A, Chapman, S.K. | | Deposit date: | 2006-10-24 | | Release date: | 2007-09-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Q403K mutant heme domain of flavocytochrome P450 BM3
To be published
|
|
1B7Q
 
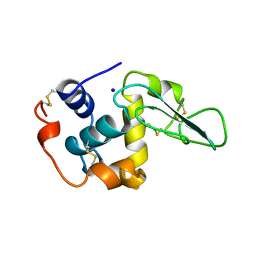 | | VERIFICATION OF SPMP USING MUTANT HUMAN LYSOZYMES | | Descriptor: | PROTEIN (LYSOZYME), SODIUM ION | | Authors: | Takano, K, Ota, M, Ogasahara, K, Yamagata, Y, Nishikawa, K, Yutani, K. | | Deposit date: | 1999-01-25 | | Release date: | 1999-02-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Experimental verification of the 'stability profile of mutant protein' (SPMP) data using mutant human lysozymes.
Protein Eng., 12, 1999
|
|
7F4Z
 
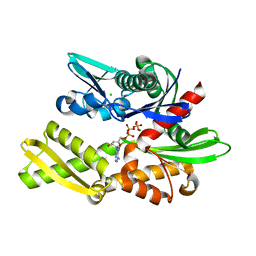 | | X-ray crystal structure of Y149A mutated Hsp72-NBD in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, Heat shock 70 kDa protein 1B, ... | | Authors: | Yokoyama, T, Fujii, S, Nabeshima, Y, Mizuguchi, M. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Neutron crystallographic analysis of the nucleotide-binding domain of Hsp72 in complex with ADP.
Iucrj, 9, 2022
|
|
3MN7
 
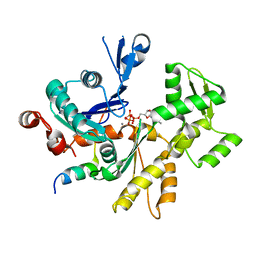 | | Structures of actin-bound WH2 domains of Spire and the implication for filament nucleation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin-5C, CALCIUM ION, ... | | Authors: | Ducka, A.M, Sitar, T, Popowicz, G.M, Huber, R, Holak, T.A. | | Deposit date: | 2010-04-21 | | Release date: | 2010-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of actin-bound Wiskott-Aldrich syndrome protein homology 2 (WH2) domains of Spire and the implication for filament nucleation.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
7F2X
 
 | | Crystal structure of MEK1 C121S mutant | | Descriptor: | MEK1 F11, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Fujioka, Y, Noda, N.N. | | Deposit date: | 2021-06-15 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.007 Å) | | Cite: | Qualitative differences in disease-associated MEK mutants reveal molecular signatures and aberrant signaling-crosstalk in cancer.
Nat Commun, 13, 2022
|
|
3MND
 
 | |
7F4X
 
 | | Joint neutron and X-ray crystal structure of the nucleotide-binding domain of Hsp72 in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Heat shock 70 kDa protein 1B, MAGNESIUM ION, ... | | Authors: | Yokoyama, T, Ostermann, A, Schrader, T.E. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-29 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (1.6 Å), X-RAY DIFFRACTION | | Cite: | Neutron crystallographic analysis of the nucleotide-binding domain of Hsp72 in complex with ADP.
Iucrj, 9, 2022
|
|
