7Z3Z
 
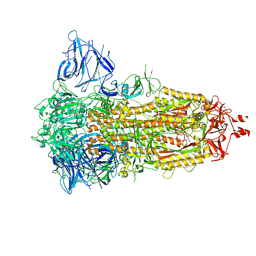 | | Locked Wuhan SARS-CoV2 Prefusion Spike ectodomain with lipid bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, STEARIC ACID, ... | | Authors: | Duyvesteyn, H.M.E, Carrique, L, Ren, J, Stuart, D.I, Fry, E.E. | | Deposit date: | 2022-03-03 | | Release date: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The SARS-CoV-2 Spike harbours a lipid binding pocket which modulates stability of the prefusion trimer
bioRxiv, 2020
|
|
7NCO
 
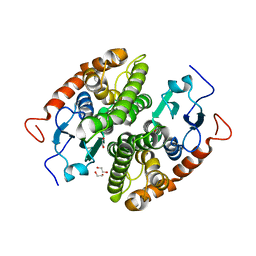 | | Glutathione-S-transferase GliG mutant K127R | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Glutathione S-transferase GliG | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2021-01-29 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Mechanistic Insights into C-S Bond Formation in Gliotoxin.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
5LRW
 
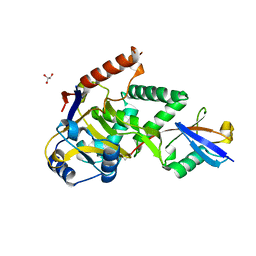 | | Structure of Cezanne/OTUD7B OTU domain bound to ubiquitin | | Descriptor: | GLYCEROL, OTU domain-containing protein 7B, Polyubiquitin-B | | Authors: | Mevissen, T.E.T, Kulathu, Y, Mulder, M.P.C, Geurink, P.P, Maslen, S.L, Gersch, M, Elliott, P.R, Burke, J.E, van Tol, B.D.M, Akutsu, M, El Oualid, F, Kawasaki, M, Freund, S.M.V, Ovaa, H, Komander, D. | | Deposit date: | 2016-08-22 | | Release date: | 2016-10-19 | | Last modified: | 2017-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis of Lys11-polyubiquitin specificity in the deubiquitinase Cezanne.
Nature, 538, 2016
|
|
7ZDQ
 
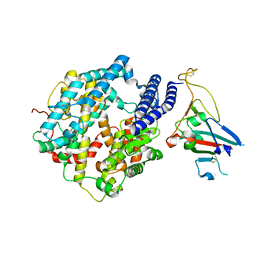 | | Cryo-EM structure of Human ACE2 bound to a high-affinity SARS CoV-2 mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike protein S1 | | Authors: | Bate, N, Savva, C.G, Moody, P.C.E, Brown, E.A, Schwabe, W.R, Brindle, N.P.J, Ball, J.K, Sale, J.E. | | Deposit date: | 2022-03-29 | | Release date: | 2022-05-18 | | Last modified: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | In vitro evolution predicts emerging SARS-CoV-2 mutations with high affinity for ACE2 and cross-species binding.
Plos Pathog., 18, 2022
|
|
5LRN
 
 | | Structure of mono-zinc MCR-1 in P21 space group | | Descriptor: | GLYCEROL, Phosphatidylethanolamine transferase Mcr-1, ZINC ION | | Authors: | Hinchliffe, P, Paterson, N.G, Spencer, J. | | Deposit date: | 2016-08-19 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Insights into the Mechanistic Basis of Plasmid-Mediated Colistin Resistance from Crystal Structures of the Catalytic Domain of MCR-1.
Sci Rep, 7, 2017
|
|
1F3W
 
 | | RECOMBINANT RABBIT MUSCLE PYRUVATE KINASE | | Descriptor: | MANGANESE (II) ION, POTASSIUM ION, PYRUVATE KINASE, ... | | Authors: | Wooll, J.O, Friesen, R.H.E, White, M.A, Watowich, S.J, Fox, R.O, Lee, J.C, Czerwinski, E.W. | | Deposit date: | 2000-06-06 | | Release date: | 2001-10-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and functional linkages between subunit interfaces in mammalian pyruvate kinase.
J.Mol.Biol., 312, 2001
|
|
6O8W
 
 | |
4XRY
 
 | | Human Cytochrome P450 2D6 BACE1 Inhibitor 5 Complex | | Descriptor: | (4aR,6R,8aS)-8a-(2,4-difluorophenyl)-6-(1-methyl-1H-pyrazol-4-yl)-4,4a,5,6,8,8a-hexahydropyrano[3,4-d][1,3]thiazin-2-amine, Cytochrome P450 2D6, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Johnson, E.F, Fan, Y. | | Deposit date: | 2015-01-21 | | Release date: | 2015-05-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Utilizing Structures of CYP2D6 and BACE1 Complexes To Reduce Risk of Drug-Drug Interactions with a Novel Series of Centrally Efficacious BACE1 Inhibitors.
J.Med.Chem., 58, 2015
|
|
8PDI
 
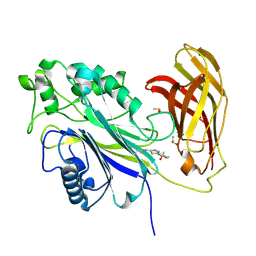 | | The phosphatase and C2 domains of SHIP1 with covalent Z1763271112 | | Descriptor: | (5-phenyl-1,3,4-thiadiazol-2-yl)methanimine, DIMETHYL SULFOXIDE, Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 1 | | Authors: | Bradshaw, W.J, Moreira, T, Pascoa, T.C, Bountra, C, Chalk, R, von Delft, F, Brennan, P.E, Gileadi, O. | | Deposit date: | 2023-06-12 | | Release date: | 2023-07-26 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Regulation of inositol 5-phosphatase activity by the C2 domain of SHIP1 and SHIP2.
Structure, 32, 2024
|
|
6X8M
 
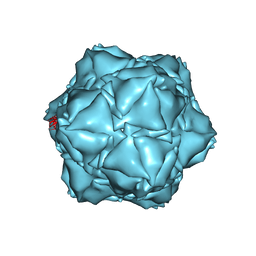 | | CryoEM structure of the holo-SrpI encapsulin complex from Synechococcus elongatus PCC 7942 | | Descriptor: | Protein SrpI | | Authors: | LaFrance, B.J, Nichols, R.J, Phillips, N.R, Oltrogge, L.M, Valentin-Alvarado, L.E, Bischoff, A.J, Savage, D.F, Nogales, E. | | Deposit date: | 2020-06-01 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Discovery and characterization of a novel family of prokaryotic nanocompartments involved in sulfur metabolism.
Elife, 10, 2021
|
|
3MVA
 
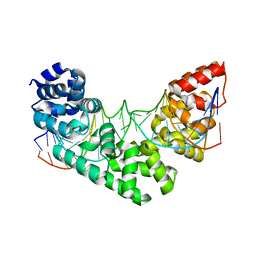 | | Crystal structure of human MTERF1 bound to the termination sequence | | Descriptor: | 5'-D(*AP*TP*TP*AP*CP*CP*GP*GP*GP*CP*TP*CP*TP*GP*CP*CP*AP*TP*CP*TP*TP*A)-3'), 5'-D(*TP*AP*AP*GP*AP*TP*GP*GP*CP*AP*GP*AP*GP*CP*CP*CP*GP*GP*TP*AP*AP*T)-3'), Transcription termination factor, ... | | Authors: | Yakubovskaya, E, Mejia, E, Byrnes, J, Hambardjieva, E, Garcia-Diaz, M. | | Deposit date: | 2010-05-03 | | Release date: | 2010-06-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Helix unwinding and base flipping enable human MTERF1 to terminate mitochondrial transcription.
Cell(Cambridge,Mass.), 141, 2010
|
|
6U96
 
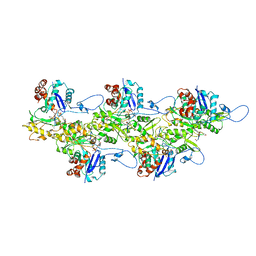 | | Actin phalloidin at BeFx state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Das, S, Ge, P, Durer, Z.A.O, Grintsevich, E.E, Zhou, Z.H, Reisler, E. | | Deposit date: | 2019-09-06 | | Release date: | 2020-05-13 | | Last modified: | 2020-05-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | D-loop Dynamics and Near-Atomic-Resolution Cryo-EM Structure of Phalloidin-Bound F-Actin.
Structure, 28, 2020
|
|
7NLY
 
 | | Crystal structure of Mycobacterium tuberculosis ArgB in complex with 2-Chlorobenzimidazole. | | Descriptor: | 2-chloranyl-1~{H}-benzimidazole, Acetylglutamate kinase, SULFATE ION | | Authors: | Mendes, V, Thomas, S.E, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2021-02-22 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.246 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7NLT
 
 | | Crystal structure of Mycobacterium tuberculosis ArgB in complex with 4-(4-methylpiperazin-1-yl)benzoic acid | | Descriptor: | 4-(4-methylpiperazin-1-yl)benzoic acid, Acetylglutamate kinase, SULFATE ION | | Authors: | Mendes, V, Thomas, S.E, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2021-02-22 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.225 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7NNB
 
 | | Crystal structure of Mycobacterium tuberculosis ArgB in complex with 2,8-bis(trifluoromethyl)quinolin-4-ol. | | Descriptor: | 1,2-ETHANEDIOL, 2,8-bis(trifluoromethyl)quinolin-4-ol, Acetylglutamate kinase, ... | | Authors: | Mendes, V, Thomas, S.E, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2021-02-24 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.186 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7ZA2
 
 | | GPC3-Unc5D octamer structure and role in cell migration | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glypican-3, ... | | Authors: | Akkermans, O, Delloye-Bourgeois, C, Peregrina, C, Carrasquero, M, Kokolaki, M, Berbeira-Santana, M, Chavent, M, Reynaud, F, Ritu, R, Agirre, J, Aksu, M, White, E, Lowe, E, Ben Amar, D, Zaballa, S, Huo, J, Pakos, I, McCubbin, P, Comoletti, D, Owens, R, Robinson, C, Castellani, V, del Toro, D, Seiradake, E. | | Deposit date: | 2022-03-21 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (4.6 Å) | | Cite: | GPC3-Unc5 receptor complex structure and role in cell migration.
Cell, 185, 2022
|
|
7NLR
 
 | | Crystal structure of Mycobacterium tuberculosis ArgB in complex with 2-phenyl-1H-imidazole | | Descriptor: | 2-phenyl-1H-imidazole, Acetylglutamate kinase, SULFATE ION | | Authors: | Mendes, V, Thomas, S.E, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2021-02-22 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.254 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7NN8
 
 | | Crystal structure of Mycobacterium tuberculosis ArgB in complex with 1H-indole-3-carbonitrile. | | Descriptor: | 1,2-ETHANEDIOL, 1~{H}-indole-3-carbonitrile, Acetylglutamate kinase, ... | | Authors: | Mendes, V, Thomas, S.E, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2021-02-24 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.267 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7NLQ
 
 | | Crystal structure of Mycobacterium tuberculosis ArgB in complex with 2-(isoxazol-5-yl)phenol | | Descriptor: | 2-(1,2-oxazol-5-yl)phenol, Acetylglutamate kinase, SULFATE ION | | Authors: | Mendes, V, Thomas, S.E, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2021-02-22 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.497 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7NLN
 
 | | Crystal structure of Mycobacterium tuberculosis ArgB in complex with N-acetyl-glutamate | | Descriptor: | 1,2-ETHANEDIOL, Acetylglutamate kinase, N-ACETYL-L-GLUTAMATE | | Authors: | Mendes, V, Thomas, S.E, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2021-02-22 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
6RLW
 
 | |
6UEU
 
 | |
7A4O
 
 | | Structure of DYRK1A in complex with AMPNP | | Descriptor: | Dual specificity tyrosine-phosphorylation-regulated kinase 1A, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Dokurno, P, Surgenor, A.E, Hubbard, R.E. | | Deposit date: | 2020-08-20 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fragment-Derived Selective Inhibitors of Dual-Specificity Kinases DYRK1A and DYRK1B.
J.Med.Chem., 64, 2021
|
|
7NLO
 
 | | Crystal structure of Mycobacterium tuberculosis ArgB in complex with L-arginine | | Descriptor: | 1,2-ETHANEDIOL, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, ARGININE, ... | | Authors: | Mendes, V, Thomas, S.E, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2021-02-22 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7NLZ
 
 | | Crystal structure of Mycobacterium tuberculosis ArgB in complex with 5-Methoxy-6-(trifluoromethyl)indole. | | Descriptor: | 5-methoxy-6-(trifluoromethyl)-1~{H}-indole, Acetylglutamate kinase, SULFATE ION | | Authors: | Mendes, V, Thomas, S.E, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2021-02-22 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.163 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
