4EVF
 
 | |
2XSM
 
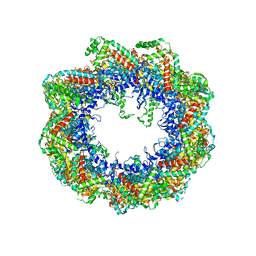 | | Crystal structure of the mammalian cytosolic chaperonin CCT in complex with tubulin | | Descriptor: | CCT | | Authors: | Munoz, I.G, Yebenes, H, Zhou, M, Mesa, P, Serna, M, Bragado-Nilsson, E, Beloso, A, Robinson, C.V, Valpuesta, J.M, Montoya, G. | | Deposit date: | 2010-10-29 | | Release date: | 2010-12-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (5.5 Å) | | Cite: | Crystal Structure of the Open Conformation of the Mammalian Chaperonin Cct in Complex with Tubulin.
Nat.Struct.Mol.Biol., 18, 2011
|
|
4EL1
 
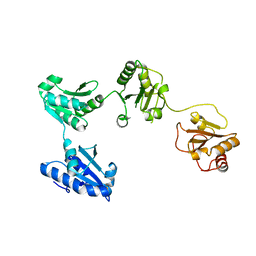 | | Crystal structure of oxidized hPDI (abb'xa') | | Descriptor: | Protein disulfide-isomerase | | Authors: | Wang, C, Li, W, Ren, J, Ke, H, Gong, W, Feng, W, Wang, C.-C. | | Deposit date: | 2012-04-10 | | Release date: | 2013-04-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.883 Å) | | Cite: | Structural insights into the redox-regulated dynamic conformations of human protein disulfide isomerase
Antioxid Redox Signal, 19, 2013
|
|
4EKZ
 
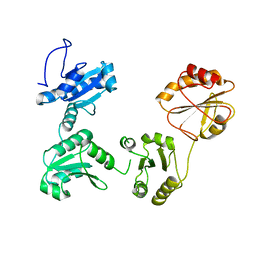 | | Crystal structure of reduced hPDI (abb'xa') | | Descriptor: | Protein disulfide-isomerase | | Authors: | Wang, C, Li, W, Ren, J, Ke, H, Gong, W, Feng, W, Wang, C.-C. | | Deposit date: | 2012-04-10 | | Release date: | 2013-04-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural insights into the redox-regulated dynamic conformations of human protein disulfide isomerase
Antioxid Redox Signal, 19, 2013
|
|
4TXK
 
 | |
6O9L
 
 | | Human holo-PIC in the closed state | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Yan, C.L, Dodd, T, He, Y, Tainer, J.A, Tsutakawa, S.E, Ivanov, I. | | Deposit date: | 2019-03-14 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Transcription preinitiation complex structure and dynamics provide insight into genetic diseases.
Nat.Struct.Mol.Biol., 26, 2019
|
|
4UI4
 
 | | Crystal structure of human tankyrase 2 in complex with TA-29 | | Descriptor: | BENZYL N-{[4-(4-OXO-3,4-DIHYDROQUINAZOLIN-2-YL)PHENYL]METHYL}CARBAMATE, SULFATE ION, TANKYRASE-2, ... | | Authors: | Haikarainen, T, Lehtio, L. | | Deposit date: | 2015-03-27 | | Release date: | 2016-04-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-Activity Relationships of 2-Arylquinazolin-4-Ones as Highly Selective and Potent Inhibitors of the Tankyrases.
Eur.J.Med.Chem., 118, 2016
|
|
4UDP
 
 | | Crystal structure of 5-hydroxymethylfurfural oxidase (HMFO) in the oxidized state | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLUCOSE-METHANOL-CHOLINE OXIDOREDUCTASE | | Authors: | Dijkman, W, Binda, C, Fraaije, M, Mattevi, A. | | Deposit date: | 2014-12-11 | | Release date: | 2015-03-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Based Enzyme Tailoring of 5-Hydroxymethylfurfural Oxidase
Acs Catalysis, 5, 2015
|
|
4UDR
 
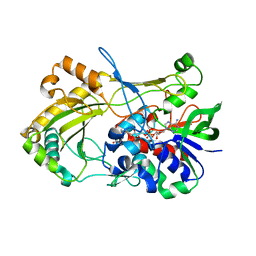 | | Crystal structure of the H467A mutant of 5-hydroxymethylfurfural oxidase (HMFO) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, FORMIC ACID, GLUCOSE-METHANOL-CHOLINE OXIDOREDUCTASE | | Authors: | Dijkman, W, Binda, C, Fraaije, M, Mattevi, A. | | Deposit date: | 2014-12-11 | | Release date: | 2015-03-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Based Enzyme Tailoring of 5-Hydroxymethylfurfural Oxidase
Acs Catalysis, 5, 2015
|
|
4UDQ
 
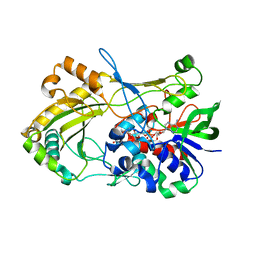 | | Crystal structure of 5-hydroxymethylfurfural oxidase (HMFO) in the reduced state | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, FORMIC ACID, GLUCOSE-METHANOL-CHOLINE OXIDOREDUCTASE | | Authors: | Dijkman, W, Binda, C, Fraaije, M, Mattevi, A. | | Deposit date: | 2014-12-11 | | Release date: | 2015-03-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Based Enzyme Tailoring of 5-Hydroxymethylfurfural Oxidase
Acs Catalysis, 5, 2015
|
|
1OSV
 
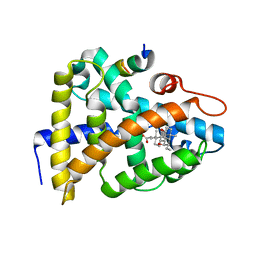 | | STRUCTURAL BASIS FOR BILE ACID BINDING AND ACTIVATION OF THE NUCLEAR RECEPTOR FXR | | Descriptor: | 6-ETHYL-CHENODEOXYCHOLIC ACID, Bile acid receptor, Nuclear receptor coactivator 2 | | Authors: | Mi, L.Z, Devarakonda, S, Harp, J.M, Han, Q, Pellicciari, R, Willson, T.M, Khorasanizadeh, S, Rastinejad, F. | | Deposit date: | 2003-03-20 | | Release date: | 2004-03-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Bile Acid Binding and Activation of the Nuclear Receptor FXR
Mol.Cell, 11, 2003
|
|
1UV4
 
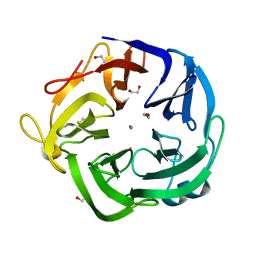 | | Native Bacillus subtilis Arabinanase Arb43A | | Descriptor: | 1,2-ETHANEDIOL, ARABINAN-ENDO 1,5-ALPHA-L-ARABINASE, CALCIUM ION | | Authors: | Nurizzo, D, Taylor, E.J, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2004-01-14 | | Release date: | 2005-02-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Tailored Catalysts for Plant Cell-Wall Degradation: Redesigning the Exo/Endo Preference of Cellvibrio Japonicus Arabinanase 43A
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
8YBX
 
 | |
9EOF
 
 | | Structure of the human INTS5/8/10/15 subcomplex | | Descriptor: | Integrator complex subunit 10, Integrator complex subunit 15, Integrator complex subunit 5, ... | | Authors: | Razew, M, Galej, W.P. | | Deposit date: | 2024-03-14 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Structural basis of the Integrator complex assembly and association with transcription factors.
Mol.Cell, 2024
|
|
9EP4
 
 | | Structure of Integrator subcomplex INTS5/8/15 | | Descriptor: | Integrator complex subunit 15, Integrator complex subunit 5, Integrator complex subunit 8 | | Authors: | Razew, M, Galej, W.P. | | Deposit date: | 2024-03-17 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of the Integrator complex assembly and association with transcription factors.
Mol.Cell, 2024
|
|
9FA4
 
 | |
9BRB
 
 | |
9BRD
 
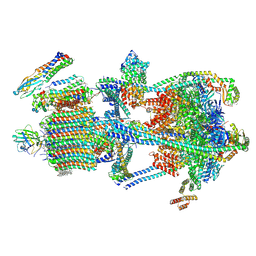 | | Synaptic Vesicle V-ATPase with synaptophysin and SidK, State 3 | | Descriptor: | (7R)-4,7-DIHYDROXY-N,N,N-TRIMETHYL-10-OXO-3,5,9-TRIOXA-4-PHOSPHAHEPTACOSAN-1-AMINIUM 4-OXIDE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Coupland, C.E, Rubinstein, J.L. | | Deposit date: | 2024-05-11 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | High-resolution electron cryomicroscopy of V-ATPase in native synaptic vesicles.
Science, 2024
|
|
8RJW
 
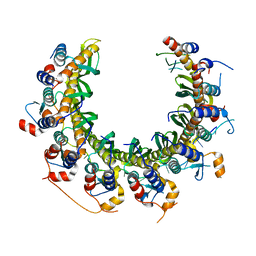 | | Human RAD52 open ring - ssDNA complex | | Descriptor: | DNA repair protein RAD52 homolog, MAGNESIUM ION, ssDNA | | Authors: | Liang, C.C, West, S.C. | | Deposit date: | 2023-12-21 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Mechanism of single-stranded DNA annealing by RAD52-RPA complex.
Nature, 629, 2024
|
|
1WAL
 
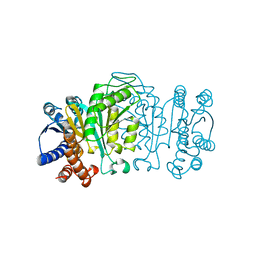 | | 3-ISOPROPYLMALATE DEHYDROGENASE (IPMDH) MUTANT (M219A)FROM THERMUS THERMOPHILUS | | Descriptor: | PROTEIN (3-ISOPROPYLMALATE DEHYDROGENASE) | | Authors: | Wallon, G, Kryger, G, Lovett, S.T, Oshima, T, Ringe, D, Petsko, G.A. | | Deposit date: | 1999-05-17 | | Release date: | 1999-05-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal structures of Escherichia coli and Salmonella typhimurium 3-isopropylmalate dehydrogenase and comparison with their thermophilic counterpart from Thermus thermophilus.
J.Mol.Biol., 266, 1997
|
|
4V4F
 
 | | The structure of the trp RNA-binding attenuation protein (TRAP) bound to a RNA molecule containing UAGAU repeats | | Descriptor: | 5'-R(*UP*AP*GP*AP*UP)-3', TRANSCRIPTION ATTENUATION PROTEIN MTRB, TRYPTOPHAN | | Authors: | Hopcroft, N.H, Manfredo, A, Wendt, A.L, Brzozowski, A.M, Gollnick, P, Antson, A.A. | | Deposit date: | 2003-12-08 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Interaction of RNA with Trap: The Role of Triplet Repeats and Separating Spacer Nucleotides
J.Mol.Biol., 338, 2004
|
|
9B8O
 
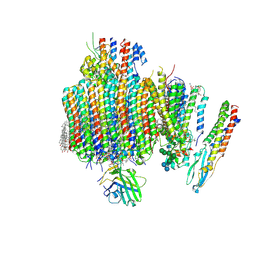 | | Synaptic Vesicle V-ATPase with synaptophysin and SidK, State 3, Vo | | Descriptor: | (7R)-4,7-DIHYDROXY-N,N,N-TRIMETHYL-10-OXO-3,5,9-TRIOXA-4-PHOSPHAHEPTACOSAN-1-AMINIUM 4-OXIDE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Coupland, C.E, Rubinstein, J.L. | | Deposit date: | 2024-03-31 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | High-resolution electron cryomicroscopy of V-ATPase in native synaptic vesicles.
Science, 2024
|
|
9EOC
 
 | |
9FA7
 
 | |
8Z50
 
 | | Crystal structure of the ASF1-H3T-H4 complex | | Descriptor: | Histone H3.1t, Histone H4, Histone chaperone ASF1A | | Authors: | Xu, L. | | Deposit date: | 2024-04-18 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into instability of the nucleosome driven by histone variant H3T.
Biochem.Biophys.Res.Commun., 727, 2024
|
|
