2L55
 
 | |
9EZZ
 
 | |
2LGE
 
 | |
7BH8
 
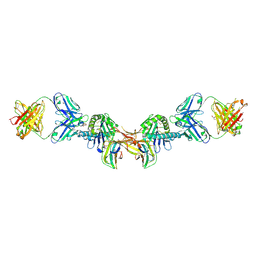 | | 3H4-Fab HLA-E-VL9 co-complex | | Descriptor: | 3H4 Fab heavy chain, 3H4 Fab light chain, Beta-2-microglobulin, ... | | Authors: | Walters, L.C, Rozbesky, D. | | Deposit date: | 2021-01-10 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mouse and human antibodies bind HLA-E-leader peptide complexes and enhance NK cell cytotoxicity.
Commun Biol, 5, 2022
|
|
9F0E
 
 | |
9C4I
 
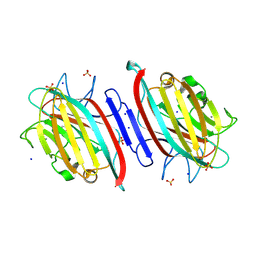 | | Centrolobium microchaete seed lectin (CML) complexed with Man1-3Man-OMe | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Nascimento, K.S, Pinto-Junior, V.R, Lima, F.E.O, Osterne, V.J.S, Oliveira, M.V, Ferreira, V.M.S, Cavada, B.S. | | Deposit date: | 2024-06-04 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Centrolobium microchaete seed lectin (CML) complexed with Man1-3Man-OMe
To Be Published
|
|
2LCZ
 
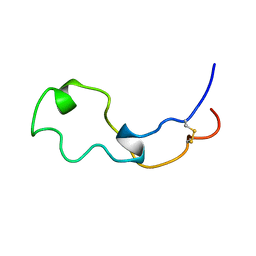 | |
9BRW
 
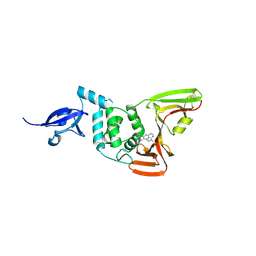 | | SARS-CoV-2 Papain-like Protease (PLpro) with Fragment 7 | | Descriptor: | CHLORIDE ION, N-(2-chlorophenyl)-1-methyl-1H-pyrazolo[3,4-d]pyrimidin-4-amine, Papain-like protease nsp3, ... | | Authors: | Amporndanai, K, Zhao, B, Fesik, S.W. | | Deposit date: | 2024-05-11 | | Release date: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Fragment-Based Screen of SARS-CoV-2 Papain-like Protease (PLpro)
Acs Med.Chem.Lett., 2024
|
|
9BGR
 
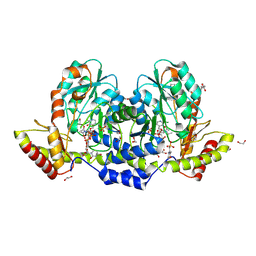 | | X-ray structure of the aminotransferase from Vibrio vulnificus responsible for the biosynthesis of 2,3-diacetamido-4-amino-2,3,4-trideoxy-arabinose in the presence of its external aldimine with 2,3-diacetamido-4-amino-2,3,4-trideoxy-l-arabinose | | Descriptor: | (2R,3R,4R,5R)-3,4-diacetamido-5-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methoxy)oxan-2-yl [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxyoxolan-2-yl]methyl dihydrogen diphosphate (non-preferred name), 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Fait, D.J, Thoden, J.B, Holden, H.M. | | Deposit date: | 2024-04-19 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Biochemical Investigation of an Aminotransferase Required for the Production of 2,3,4-triacetamido-2,3,4-trideoxy-L-arabinose
To Be Published
|
|
2LIU
 
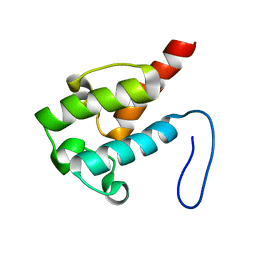 | | NMR structure of holo-ACPI domain from CurA module from Lyngbya majuscula | | Descriptor: | CurA | | Authors: | Busche, A.E, Gottstein, D, Hein, C, Ripin, N, Pader, I, Tufar, P, Eisman, E.B, Gu, L, Walsh, C.T, Loehr, F, Sherman, D.H, Guntert, P, Dotsch, V. | | Deposit date: | 2011-09-01 | | Release date: | 2011-12-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Characterization of Molecular Interactions between ACP and Halogenase Domains in the Curacin A Polyketide Synthase.
Acs Chem.Biol., 7, 2012
|
|
2LF8
 
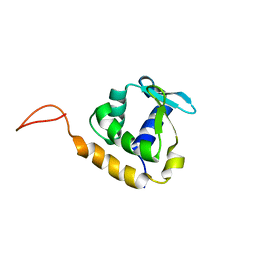 | |
2LDA
 
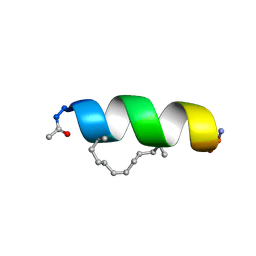 | | Solution structure of the estrogen receptor-binding stapled peptide SP2 (Ac-HKXLHQXLQDS-NH2) | | Descriptor: | Estrogen receptor-binding stapled peptide SP2 | | Authors: | Phillips, C, Bazin, R, Bent, A, Davies, N, Moore, R, Pannifer, A, Pickford, A, Prior, S, Read, C, Roberts, L, Schade, M, Scott, A, Brown, D, Xu, B, Irving, S. | | Deposit date: | 2011-05-20 | | Release date: | 2011-07-06 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Design and structure of stapled peptides binding to estrogen receptors.
J.Am.Chem.Soc., 133, 2011
|
|
4U13
 
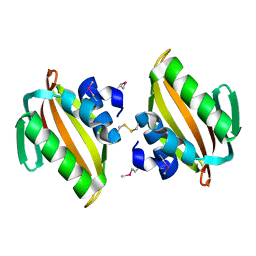 | | Crystal structure of putative polyketide cyclase (protein SMa1630) from Sinorhizobium meliloti at 2.3 A resolution | | Descriptor: | putative polyketide cyclase SMa1630 | | Authors: | Shabalin, I.G, Bacal, P, Osinski, T, Cooper, D.R, Szlachta, K, Stead, M, Grabowski, M, Hammonds, J, Ahmed, M, Hillerich, B.S, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-07-14 | | Release date: | 2014-09-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a putative polyketide cyclase (protein SMa1630) from Sinorhizobium meliloti at 2.3 A resolution
to be published
|
|
2LQX
 
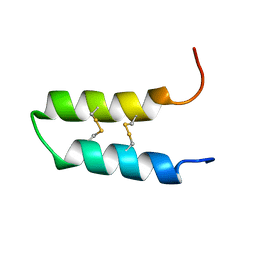 | | NMR spatial structure of the trypsin inhibitor BWI-2c from the buckwheat seeds | | Descriptor: | Trypsin inhibitor BWI-2c | | Authors: | Mineev, K.S, Vassilevski, A.A, Oparin, P.B, Grishin, E.V, Egorov, T.A. | | Deposit date: | 2012-03-19 | | Release date: | 2012-05-30 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Buckwheat trypsin inhibitor with helical hairpin structure belongs to a new family of plant defence peptides.
Biochem.J., 446, 2012
|
|
2LG7
 
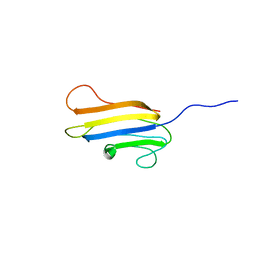 | |
2LNB
 
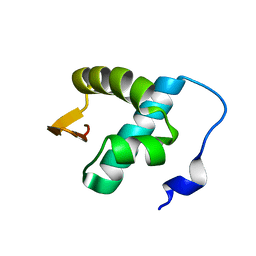 | | Solution NMR structure of N-terminal domain (6-74) of human ZBP1 protein, Northeast Structural Genomics Consortium Target HR8174A. | | Descriptor: | Z-DNA-binding protein 1 | | Authors: | Yang, Y, Ramelot, T.A, Hamilton, K, Kohan, E, Wang, D, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-12-20 | | Release date: | 2012-01-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of N-terminal domain (6-74) of human ZBP1 protein, Northeast Structural Genomics Consortium Target HR8174A
To be Published
|
|
2L5Q
 
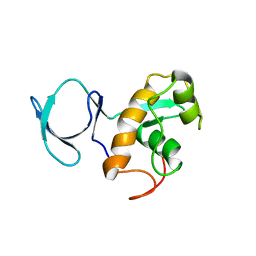 | | Solution NMR Structure of BVU_3817 from Bacteroides vulgatus, Northeast Structural Genomics Consortium Target BvR159 | | Descriptor: | Uncharacterized protein | | Authors: | Mills, J.L, Eletsky, A, Lee, H, Wang, H, Ciccosanti, C, Hamilton, K, Acton, T.B, Xiao, R, Everett, J.K, Prestegard, J.H, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-11-03 | | Release date: | 2011-01-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Northeast Structural Genomics Consortium Target BvR159
To be Published
|
|
2LTM
 
 | | Solution NMR Structure of NFU1 Iron-Sulfur Cluster Scaffold Homolog from Homo sapiens, Northeast Structural Genomics Consortium (NESG) Target HR2876B | | Descriptor: | NFU1 iron-sulfur cluster scaffold homolog, mitochondrial | | Authors: | Liu, G, Xiao, R, Janjua, H, Hamilton, K, Shastry, R, Kohan, E, Acton, T.B, Everett, J.K, Lee, H, Huang, Y.J, Montelione, G.T, Northeast Structural Genomics Consortium (NESG), Mitochondrial Protein Partnership (MPP) | | Deposit date: | 2012-05-29 | | Release date: | 2012-07-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of NFU1 Iron-Sulfur Cluster Scaffold Homolog from Homo sapiens, Northeast Structural Genomics Consortium (NESG) Target HR2876B
To be Published
|
|
2L39
 
 | | Mouse prion protein fragment 121-231 AT 37 C | | Descriptor: | Major prion protein | | Authors: | Christen, B, Damberger, F.F, Perez, D.R, Hornemann, S, Wuthrich, K. | | Deposit date: | 2010-09-10 | | Release date: | 2011-08-10 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Cellular prion protein conformation and function.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2LTL
 
 | | Solution NMR Structure of NifU-like protein from Saccharomyces cerevisiae, Northeast Structural Genomics Consortium (NESG) Target YR313A | | Descriptor: | NifU-like protein, mitochondrial | | Authors: | Liu, G, Xiao, R, Hamilton, K, Janjua, H, Shastry, R, Kohan, E, Acton, T.B, Everett, J.K, Lee, H, Huang, Y.J, Montelione, G.T, Northeast Structural Genomics Consortium (NESG), Mitochondrial Protein Partnership (MPP) | | Deposit date: | 2012-05-29 | | Release date: | 2012-07-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of NifU-like protein from Saccharomyces cerevisiae, Northeast Structural Genomics Consortium (NESG) Target YR313A
To be Published
|
|
2LD8
 
 | |
2LRC
 
 | |
2LU9
 
 | |
2LU1
 
 | | pfsub2 solution NMR structure | | Descriptor: | Subtilase | | Authors: | He, Y, Chen, Y, Ruan, B, O'Brochta, D, Bryan, P, Orban, J. | | Deposit date: | 2012-06-06 | | Release date: | 2012-10-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of a sheddase inhibitor prodomain from the malarial parasite Plasmodium falciparum.
Proteins, 80, 2012
|
|
2MB2
 
 | | parallel-stranded G-quadruplex in DNA poly-G stretches | | Descriptor: | DNA_(5'-D(*TP*TP*GP*GP*GP*GP*GP*GP*GP*GP*GP*GP*GP*GP*GP*GP*GP*T)-3') | | Authors: | Sengar, A, Heddi, B, Phan, A.T. | | Deposit date: | 2013-07-24 | | Release date: | 2014-12-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Formation of g-quadruplexes in poly-g sequences: structure of a propeller-type parallel-stranded g-quadruplex formed by a g15 stretch.
Biochemistry, 53, 2014
|
|
