3DQN
 
 | |
7NZ2
 
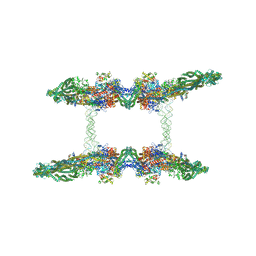 | | Cryo-EM structure of the MukBEF-MatP-DNA tetrad | | Descriptor: | 4'-PHOSPHOPANTETHEINE, ADENOSINE-5'-TRIPHOSPHATE, Acyl carrier protein, ... | | Authors: | Buermann, F, Lowe, J. | | Deposit date: | 2021-03-23 | | Release date: | 2021-07-07 | | Last modified: | 2022-06-29 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Cryo-EM structure of MukBEF reveals DNA loop entrapment at chromosomal unloading sites.
Mol.Cell, 81, 2021
|
|
4R92
 
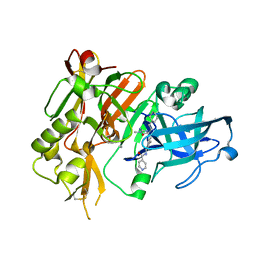 | | BACE-1 in complex with (R)-4-(2-cyclohexylethyl)-4-(((1S,3R)-3-(isonicotinamido)cyclohexyl)methyl)-1-methyl-5-oxoimidazolidin-2-iminium | | Descriptor: | Beta-secretase 1, L(+)-TARTARIC ACID, N-[(1R,3S)-3-{[(2E,4R)-4-(2-cyclohexylethyl)-2-imino-1-methyl-5-oxoimidazolidin-4-yl]methyl}cyclohexyl]pyridine-4-carboxamide | | Authors: | Orth, P, Strickland, C, Caldwell, J.P. | | Deposit date: | 2014-09-03 | | Release date: | 2014-11-05 | | Last modified: | 2014-12-17 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Discovery of potent iminoheterocycle BACE1 inhibitors.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
3E28
 
 | |
4RBV
 
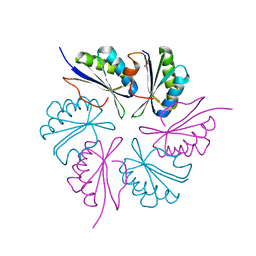 | |
4RCE
 
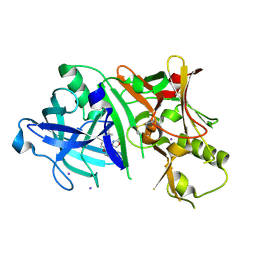 | | Crystal structure of BACE1 in complex with aminooxazoline xanthene inhibitor 2 | | Descriptor: | (4S)-2'-(2,2-dimethylpropoxy)-7'-(pyrimidin-5-yl)spiro[1,3-oxazole-4,9'-xanthen]-2-amine, Beta-secretase 1, IODIDE ION | | Authors: | Whittington, D.A, Long, A.M. | | Deposit date: | 2014-09-15 | | Release date: | 2014-12-24 | | Last modified: | 2015-01-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Lead Optimization and Modulation of hERG Activity in a Series of Aminooxazoline Xanthene beta-Site Amyloid Precursor Protein Cleaving Enzyme (BACE1) Inhibitors.
J.Med.Chem., 57, 2014
|
|
3E1G
 
 | |
3DPX
 
 | |
3DQ5
 
 | |
3DQE
 
 | |
3DR9
 
 | | Increased Distal Histidine Conformational Flexibility in the Deoxy Form of Dehaloperoxidase from Amphitrite ornata | | Descriptor: | Dehaloperoxidase A, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Chen, X, de Serrano, V.S, Betts, L, Franzen, S. | | Deposit date: | 2008-07-10 | | Release date: | 2009-01-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Distal histidine conformational flexibility in dehaloperoxidase from Amphitrite ornata.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
4REF
 
 | | Crystal Structure of TR3 LBD_L449W in complex with Molecule 2 | | Descriptor: | 1-(3,4,5-trihydroxyphenyl)hexan-1-one, GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Li, F.W, Cai, Q.X, Li, A.Z, Tian, X.Y, Wang, W.J, Yuan, W, Hou, P.P, Wu, Q, Lin, T.W. | | Deposit date: | 2014-09-22 | | Release date: | 2015-09-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Induction of Autophagic Death in Cancer Cells by Agonizing TR3 and Attenuating Akt2 Activity
Chem.Biol., 22, 2015
|
|
4REM
 
 | | Crystal structure of UDP-glucose: anthocyanidin 3-O-glucosyltransferase in complex with delphinidin | | Descriptor: | 3,5,7-trihydroxy-2-(3,4,5-trihydroxyphenyl)chromenium, GLYCEROL, UDP-glucose:anthocyanidin 3-O-glucosyltransferase | | Authors: | Hiromoto, T, Honjo, E, Tamada, T, Kuroki, R. | | Deposit date: | 2014-09-23 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural basis for acceptor-substrate recognition of UDP-glucose: anthocyanidin 3-O-glucosyltransferase from Clitoria ternatea
Protein Sci., 24, 2015
|
|
4RG3
 
 | |
3DU0
 
 | | E. coli dihydrodipicolinate synthase with first substrate, pyruvate, bound in active site | | Descriptor: | CHLORIDE ION, Dihydrodipicolinate synthase, GLYCEROL, ... | | Authors: | Dobson, R.C.J, Devenish, S.R.A, Gerrard, J.A, Jameson, G.B. | | Deposit date: | 2008-07-16 | | Release date: | 2008-11-18 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The high-resolution structure of dihydrodipicolinate synthase from Escherichia coli bound to its first substrate, pyruvate.
Acta Crystallogr.,Sect.F, 64, 2008
|
|
4RCF
 
 | | Crystal structure of BACE1 in complex with 2-aminooxazoline 4-fluoroxanthene inhibitor 49 | | Descriptor: | (4S)-2'-(3,6-dihydro-2H-pyran-4-yl)-4'-fluoro-7'-(2-fluoropyridin-3-yl)spiro[1,3-oxazole-4,9'-xanthen]-2-amine, Beta-secretase 1, GLYCEROL, ... | | Authors: | Whittington, D.A, Long, A.M. | | Deposit date: | 2014-09-15 | | Release date: | 2014-12-24 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Lead Optimization and Modulation of hERG Activity in a Series of Aminooxazoline Xanthene beta-Site Amyloid Precursor Protein Cleaving Enzyme (BACE1) Inhibitors.
J.Med.Chem., 57, 2014
|
|
4RCP
 
 | | Crystal structure of Plk1 Polo-box domain in complex with PL-2 | | Descriptor: | 1,2-ETHANEDIOL, PL-2, Serine/threonine-protein kinase PLK1 | | Authors: | Lee, W.C, Song, J.H, Kim, H.Y. | | Deposit date: | 2014-09-16 | | Release date: | 2014-11-12 | | Last modified: | 2015-01-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A new class of peptidomimetics targeting the polo-box domain of polo-like kinase 1.
J.Med.Chem., 58, 2015
|
|
4R6I
 
 | | AtxA protein, a virulence regulator from Bacillus anthracis. | | Descriptor: | Anthrax toxin expression trans-acting positive regulator, DODECYL-BETA-D-MALTOSIDE | | Authors: | Osipiuk, J, Horton, L.B, Koehler, T.M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-08-25 | | Release date: | 2014-10-22 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of Bacillus anthracis virulence regulator AtxA and effects of phosphorylated histidines on multimerization and activity.
Mol.Microbiol., 95, 2015
|
|
3DO2
 
 | |
4REL
 
 | | Crystal structure of UDP-glucose: anthocyanidin 3-O-glucosyltransferase in complex with kaempferol | | Descriptor: | 3,5,7-TRIHYDROXY-2-(4-HYDROXYPHENYL)-4H-CHROMEN-4-ONE, ACETATE ION, GLYCEROL, ... | | Authors: | Hiromoto, T, Honjo, E, Tamada, T, Kuroki, R. | | Deposit date: | 2014-09-23 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.754 Å) | | Cite: | Structural basis for acceptor-substrate recognition of UDP-glucose: anthocyanidin 3-O-glucosyltransferase from Clitoria ternatea
Protein Sci., 24, 2015
|
|
3DQ0
 
 | | Maize cytokinin oxidase/dehydrogenase complexed with N6-(3-methoxy-phenyl)adenine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cytokinin dehydrogenase 1, ... | | Authors: | Kopecny, D, Briozzo, P. | | Deposit date: | 2008-07-09 | | Release date: | 2009-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterization and biological activity of novel purine-derived inhibitor of cytokinin oxidase/dehydrogenase and its potential use for in vivo studies
To be Published
|
|
3DQJ
 
 | |
3E17
 
 | | Crystal structure of the second PDZ domain from human Zona Occludens-2 | | Descriptor: | Tight junction protein ZO-2 | | Authors: | Chen, H, Tong, S.L, Teng, M.K, Niu, L.W. | | Deposit date: | 2008-08-01 | | Release date: | 2009-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of the second PDZ domain from human zonula occludens 2
Acta Crystallogr.,Sect.F, 65, 2009
|
|
3DRU
 
 | | Crystal Structure of Gly117Phe Alpha1-Antitrypsin | | Descriptor: | Alpha-1-antitrypsin | | Authors: | Gooptu, B, Nobeli, I, Purkiss, A, Phillips, R.L, Mallya, M, Lomas, D.A, Barrett, T.E. | | Deposit date: | 2008-07-11 | | Release date: | 2009-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystallographic and cellular characterisation of two mechanisms stabilising the native fold of alpha1-antitrypsin: implications for disease and drug design.
J.Mol.Biol., 387, 2009
|
|
3DUS
 
 | | Crystal structure of SAG506-01, orthorhombic, twinned, crystal 1 | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid, Ig-like protein, MAGNESIUM ION, ... | | Authors: | Brooks, C.L, Blackler, R.J, Gerstenbruch, S, Kosma, P, Muller-Loennies, S, Brade, H, Evans, S.V. | | Deposit date: | 2008-07-17 | | Release date: | 2008-12-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Pseudo-symmetry and twinning in crystals of homologous antibody Fv fragments.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
