4N32
 
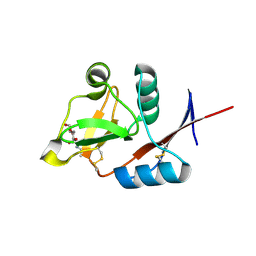 | | Structure of langerin CRD with alpha-Me-GlcNAc. | | Descriptor: | C-type lectin domain family 4 member K, CALCIUM ION, methyl 2-acetamido-2-deoxy-alpha-D-glucopyranoside | | Authors: | Feinberg, H, Rowntree, T.J.W, Tan, S.L.W, Drickamer, K, Weis, W.I, Taylor, M.E. | | Deposit date: | 2013-10-06 | | Release date: | 2013-11-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Common polymorphisms in human langerin change specificity for glycan ligands.
J.Biol.Chem., 288, 2013
|
|
1LO2
 
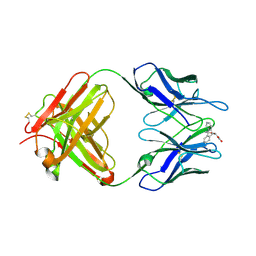 | | Retro-Diels-Alderase Catalytic Antibody | | Descriptor: | If kappa light chain, Ig gamma 2a heavy chain, [2'-CARBOXYLETHYL]-10-METHYL-ANTHRACENE ENDOPEROXIDE | | Authors: | Hugot, M, Reymond, J.L, Baumann, U. | | Deposit date: | 2002-05-06 | | Release date: | 2002-06-26 | | Last modified: | 2021-07-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A structural basis for the activity of retro-Diels-Alder catalytic antibodies: evidence for a catalytic aromatic residue.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
4N38
 
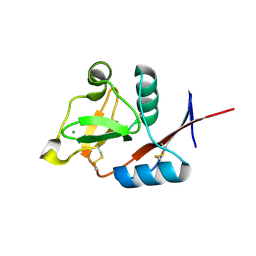 | | Structure of langerin CRD I313 D288 complexed with GlcNAc-beta1-3Gal-beta1-4GlcNAc-beta-CH2CH2N3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose, C-type lectin domain family 4 member K, CALCIUM ION, ... | | Authors: | Feinberg, H, Rowntree, T.J.W, Tan, S.L.W, Drickamer, K, Weis, W.I, Taylor, M.E. | | Deposit date: | 2013-10-06 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Common polymorphisms in human langerin change specificity for glycan ligands.
J.Biol.Chem., 288, 2013
|
|
1LO4
 
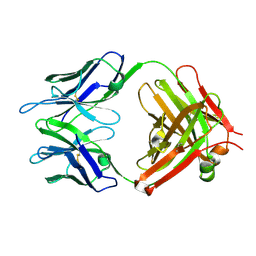 | | Retro-Diels-Alderase Catalytic antibody 9D9 | | Descriptor: | If kappa light chain, Ig gamma 2a heavy chain | | Authors: | Hugot, M, Reymond, J.L, Baumann, U. | | Deposit date: | 2002-05-06 | | Release date: | 2002-07-03 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A structural basis for the activity of retro-Diels-Alder catalytic antibodies: evidence for a catalytic aromatic residue.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1LO0
 
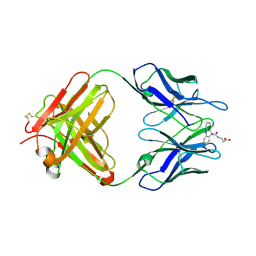 | | Catalytic Retro-Diels-Alderase Transition State Analogue Complex | | Descriptor: | 3-{[(9-CYANO-9,10-DIHYDRO-10-METHYLACRIDIN-9-YL)CARBONYL]AMINO}PROPANOIC ACID, If kappa light chain, Ig gamma 2a heavy chain | | Authors: | Hugot, M, Reymond, J.L, Baumann, U. | | Deposit date: | 2002-05-05 | | Release date: | 2002-06-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A structural basis for the activity of retro-Diels-Alder catalytic antibodies: evidence for a catalytic aromatic residue.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
5LQ6
 
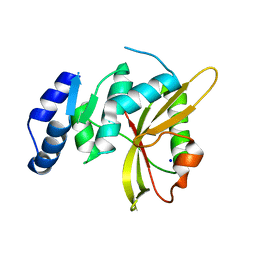 | | Salmonella effector SpvD - R161 variant | | Descriptor: | SODIUM ION, Virulence protein vsdE | | Authors: | Zhang, Y, Grabe, G.J, Rolhion, N, Yang, Y, Holden, D.W, Hare, S.A. | | Deposit date: | 2016-08-16 | | Release date: | 2016-11-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | The Salmonella Effector SpvD Is a Cysteine Hydrolase with a Serovar-specific Polymorphism Influencing Catalytic Activity, Suppression of Immune Responses, and Bacterial Virulence.
J. Biol. Chem., 291, 2016
|
|
5IV1
 
 | | Solution Structure of DNA Dodecamer with 8-oxoguanine at 4th Position | | Descriptor: | DNA (5'-D(*CP*GP*CP*(8OG)P*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Miears, H.L, Gruber, D.R, Hoppins, J.J, Kiryutin, A.S, Kasymov, R.D, Yurkovskaya, A.V, Zharkov, D.O, Smirnov, S.L. | | Deposit date: | 2016-03-18 | | Release date: | 2016-07-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | 8-Oxoguanine Affects DNA Backbone Conformation in the EcoRI Recognition Site and Inhibits Its Cleavage by the Enzyme.
Plos One, 11, 2016
|
|
5WNK
 
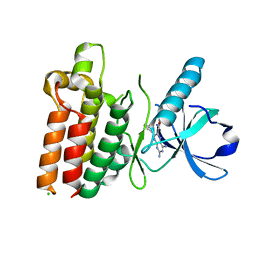 | | Crystal structure of murine receptor-interacting protein 4 (Ripk4) D143N bound to TG100-115 | | Descriptor: | 3,3'-(2,4-diaminopteridine-6,7-diyl)diphenol, CHLORIDE ION, Receptor-interacting serine/threonine-protein kinase 4 | | Authors: | Huang, C.S, Hymowitz, S.G. | | Deposit date: | 2017-08-01 | | Release date: | 2018-05-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Crystal Structure of Ripk4 Reveals Dimerization-Dependent Kinase Activity.
Structure, 26, 2018
|
|
5WNI
 
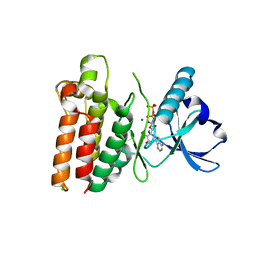 | |
5K6P
 
 | | The NMR structure of the m domain tri-helix bundle and C2 of human cardiac Myosin Binding Protein C | | Descriptor: | Myosin-binding protein C, cardiac-type | | Authors: | Michie, K.A, Kwan, A.H, Tung, C.S, Guss, J.M, Trewhella, J. | | Deposit date: | 2016-05-25 | | Release date: | 2016-11-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A Highly Conserved Yet Flexible Linker Is Part of a Polymorphic Protein-Binding Domain in Myosin-Binding Protein C.
Structure, 24, 2016
|
|
5KN7
 
 | | Lipid A secondary acyltransferase LpxM from Acinetobacter baumannii | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, GLYCEROL, Lipid A biosynthesis lauroyl acyltransferase, ... | | Authors: | Dovala, D.L, Hu, Q, Metzger IV, L.E. | | Deposit date: | 2016-06-27 | | Release date: | 2016-09-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure-guided enzymology of the lipid A acyltransferase LpxM reveals a dual activity mechanism.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5JDD
 
 | |
5JDJ
 
 | | Crystal structure of domain I10 from titin in space group P212121 | | Descriptor: | CALCIUM ION, Titin | | Authors: | Williams, R, Bogomolovas, J, Labiet, S, Mayans, O. | | Deposit date: | 2016-04-16 | | Release date: | 2016-08-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.738 Å) | | Cite: | Exploration of pathomechanisms triggered by a single-nucleotide polymorphism in titin's I-band: the cardiomyopathy-linked mutation T2580I.
Open Biology, 6, 2016
|
|
1PJZ
 
 | |
5IZP
 
 | | Solution Structure of DNA Dodecamer with 8-oxoguanine at 10th Position | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*(8OG)P*CP*G)-3') | | Authors: | Gruber, D.R, Hoppins, J.J, Miears, H.L, Kiryutin, A.S, Kasymov, R.D, Yurkovskaya, A.V, Zharkov, D.O, Smirnov, S.L. | | Deposit date: | 2016-03-25 | | Release date: | 2016-08-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | 8-Oxoguanine Affects DNA Backbone Conformation in the EcoRI Recognition Site and Inhibits Its Cleavage by the Enzyme.
Plos One, 11, 2016
|
|
5KNK
 
 | | Lipid A secondary acyltransferase LpxM from Acinetobacter baumannii with catalytic residue substitution (E127A) | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, GLYCEROL, Lipid A biosynthesis lauroyl acyltransferase, ... | | Authors: | Dovala, D.L, Hu, Q, Metzger IV, L.E. | | Deposit date: | 2016-06-28 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-guided enzymology of the lipid A acyltransferase LpxM reveals a dual activity mechanism.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1NU9
 
 | | Staphylocoagulase-Prethrombin-2 complex | | Descriptor: | IMIDAZOLE, MERCURY (II) ION, N-(sulfanylacetyl)-D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, ... | | Authors: | Friedrich, R, Bode, W, Fuentes-Prior, P, Panizzi, P, Bock, P.E. | | Deposit date: | 2003-01-31 | | Release date: | 2003-10-07 | | Last modified: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Staphylocoagulase is a prototype for the mechanism of cofactor-induced zymogen activation
NATURE, 425, 2003
|
|
5JDE
 
 | |
5WNJ
 
 | |
5ZCT
 
 | | The crystal structure of the poly-alpha-L-glutamate peptides synthetase RimK at 2.05 angstrom resolution. | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Ribosomal protein S6--L-glutamate ligase, ... | | Authors: | Arimura, Y, Kono, T, Kino, K, Kurumizaka, H. | | Deposit date: | 2018-02-20 | | Release date: | 2018-07-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural polymorphism of the Escherichia coli poly-alpha-L-glutamate synthetase RimK
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
5LQ7
 
 | | Salmonella effector SpvD - G161 variant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Przydacz, M, Grabe, G.J, Holden, D.W, Hare, S.A. | | Deposit date: | 2016-08-16 | | Release date: | 2016-11-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Salmonella Effector SpvD Is a Cysteine Hydrolase with a Serovar-specific Polymorphism Influencing Catalytic Activity, Suppression of Immune Responses, and Bacterial Virulence.
J. Biol. Chem., 291, 2016
|
|
1NU7
 
 | | Staphylocoagulase-Thrombin Complex | | Descriptor: | IMIDAZOLE, MERCURY (II) ION, N-(sulfanylacetyl)-D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, ... | | Authors: | Friedrich, R, Bode, W, Fuentes-Prior, P, Panizzi, P, Bock, P.E. | | Deposit date: | 2003-01-31 | | Release date: | 2003-10-07 | | Last modified: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Staphylocoagulase is a prototype for the mechanism of cofactor-induced zymogen activation
NATURE, 425, 2003
|
|
5WNL
 
 | |
5WA1
 
 | | CHMP4C A232T in complex with ALIX BRO1 | | Descriptor: | Charged multivesicular body protein 4c, Programmed cell death 6-interacting protein | | Authors: | Wenzel, D.M, Alam, S.L, Sundquist, W.I. | | Deposit date: | 2017-06-24 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.868 Å) | | Cite: | A cancer-associated polymorphism in ESCRT-III disrupts the abscission checkpoint and promotes genome instability.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6EJK
 
 | | Structure of a glycosyltransferase | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-alpha-D-glucopyranose, NerylNeryl pyrophosphate, Uridine-Diphosphate-Methylene-N-acetyl-galactosamine, ... | | Authors: | Ramirez, A.S, Boilevin, J, Mehdipour, A.R, Hummer, G, Darbre, T, Reymond, J.L, Locher, K.P. | | Deposit date: | 2017-09-21 | | Release date: | 2018-02-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis of the molecular ruler mechanism of a bacterial glycosyltransferase.
Nat Commun, 9, 2018
|
|
