2BI6
 
 | | NMR STUDY OF BROMELAIN INHIBITOR VI FROM PINEAPPLE STEM | | Descriptor: | BROMELAIN INHIBITOR VI | | Authors: | Hatano, K.-I. | | Deposit date: | 1995-12-07 | | Release date: | 1996-04-03 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of bromelain inhibitor IV from pineapple stem: structural similarity with Bowman-Birk trypsin/chymotrypsin inhibitor from soybean.
Biochemistry, 35, 1996
|
|
5AA3
 
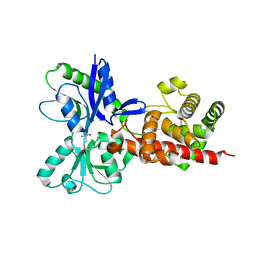 | |
5AZD
 
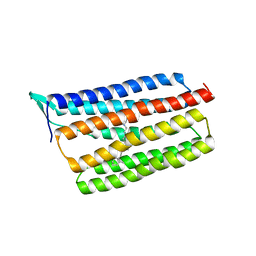 | | Crystal structure of thermophilic rhodopsin. | | Descriptor: | Bacteriorhodopsin | | Authors: | Mizutani, K, Hashimoto, N, Tsukamoto, T, Yamashita, K, Yamamoto, M, Sudo, Y, Murata, T. | | Deposit date: | 2015-09-30 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray crystallographic structure of thermophilic rhodopsin: implications for high thermal stability and optogenetic availability.
To Be Published
|
|
8WXL
 
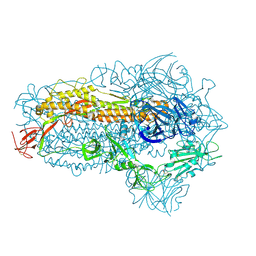 | | Structure of the SARS-CoV-2 BA.2.86 spike glycoprotein (closed state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2023-10-30 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Structural basis for receptor-binding domain mobility of the spike in SARS-CoV-2 BA.2.86 and JN.1.
Nat Commun, 15, 2024
|
|
8XUX
 
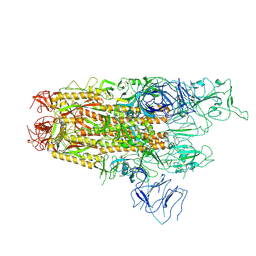 | | Structure of the SARS-CoV-2 BA.2.86 spike protein (1-up state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2024-01-14 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Structural basis for receptor-binding domain mobility of the spike in SARS-CoV-2 BA.2.86 and JN.1.
Nat Commun, 15, 2024
|
|
8XUZ
 
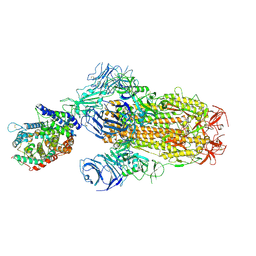 | | Structure of SARS-CoV-2 BA.2.86 spike glycoprotein in complex with ACE2 (2-up and 1-down state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2024-01-14 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structural basis for receptor-binding domain mobility of the spike in SARS-CoV-2 BA.2.86 and JN.1.
Nat Commun, 15, 2024
|
|
8XV0
 
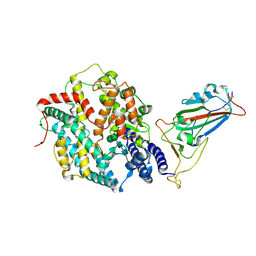 | | Structure of SARS-CoV-2 BA.2.86 spike RBD in complex with ACE2 (up state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2024-01-14 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for receptor-binding domain mobility of the spike in SARS-CoV-2 BA.2.86 and JN.1.
Nat Commun, 15, 2024
|
|
8XUY
 
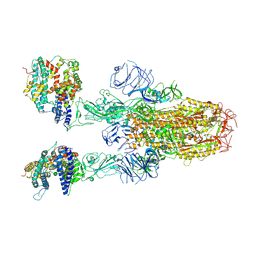 | | Structure of SARS-CoV-2 BA.2.86 spike glycoprotein in complex with ACE2 (2-up state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2024-01-14 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Structural basis for receptor-binding domain mobility of the spike in SARS-CoV-2 BA.2.86 and JN.1.
Nat Commun, 15, 2024
|
|
5AWF
 
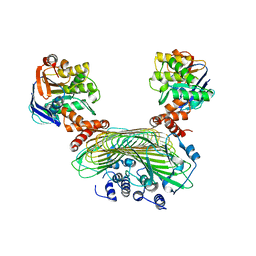 | | Crystal structure of SufB-SufC-SufD complex from Escherichia coli | | Descriptor: | FeS cluster assembly protein SufB, FeS cluster assembly protein SufD, Probable ATP-dependent transporter SufC | | Authors: | Hirabayashi, K, Wada, K. | | Deposit date: | 2015-07-03 | | Release date: | 2015-11-11 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.957 Å) | | Cite: | Functional Dynamics Revealed by the Structure of the SufBCD Complex, a Novel ATP-binding Cassette (ABC) Protein That Serves as a Scaffold for Iron-Sulfur Cluster Biogenesis
J.Biol.Chem., 290, 2015
|
|
5A5X
 
 | |
5B04
 
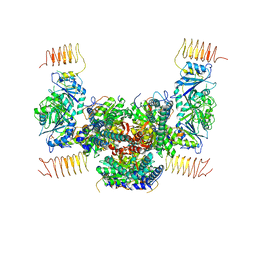 | | Crystal structure of the eukaryotic translation initiation factor 2B from Schizosaccharomyces pombe | | Descriptor: | PHOSPHATE ION, Probable translation initiation factor eIF-2B subunit beta, Probable translation initiation factor eIF-2B subunit delta, ... | | Authors: | Kashiwagi, K, Ito, T, Yokoyama, S. | | Deposit date: | 2015-10-27 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.994 Å) | | Cite: | Crystal structure of eukaryotic translation initiation factor 2B
Nature, 531, 2016
|
|
5B6F
 
 | | Crystal structure of the Fab fragment of an anti-Leukotriene C4 monoclonal antibody complexed with LTC4 | | Descriptor: | (5~{S},6~{R},7~{E},9~{E},11~{Z},14~{Z})-6-[(2~{R})-2-[[(4~{S})-4-azanyl-5-oxidanyl-5-oxidanylidene-pentanoyl]amino]-3-(2-hydroxy-2-oxoethylamino)-3-oxidanylidene-propyl]sulfanyl-5-oxidanyl-icosa-7,9,11,14-tetraenoic acid, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, SULFATE ION, ... | | Authors: | Sugahara, M, Ago, H, Saino, H, Miyano, M. | | Deposit date: | 2016-05-27 | | Release date: | 2017-05-31 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the Fab fragment of an anti-Leukotriene C4 monoclonal antibody complexed with LTC4
To Be Published
|
|
5B3V
 
 | |
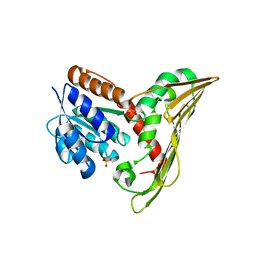
5B3U
 
 | |
5B3T
 
 | |
7VLK
 
 | | eIF2B-SFSV NSs C2-imposed | | Descriptor: | Non-structural protein NS-S, Translation initiation factor eIF-2B subunit alpha, Translation initiation factor eIF-2B subunit beta, ... | | Authors: | Kashiwagi, K, Ito, T. | | Deposit date: | 2021-10-04 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.27 Å) | | Cite: | eIF2B-capturing viral protein NSs suppresses the integrated stress response.
Nat Commun, 12, 2021
|
|
7V73
 
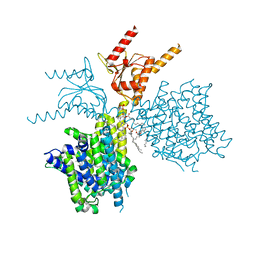 | | Thermostabilized human prestin in complex with chloride | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, CHLORIDE ION, CHOLESTEROL, ... | | Authors: | Futamata, H, Fukuda, M, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-08-21 | | Release date: | 2022-08-31 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Cryo-EM structures of thermostabilized prestin provide mechanistic insights underlying outer hair cell electromotility.
Nat Commun, 13, 2022
|
|
7V74
 
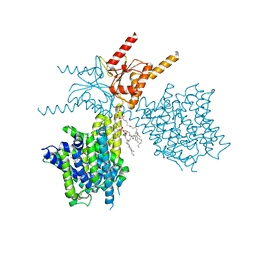 | | Thermostabilized human prestin in complex with sulfate | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, CHOLESTEROL, SULFATE ION, ... | | Authors: | Futamata, H, Fukuda, M, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-08-21 | | Release date: | 2022-08-31 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Cryo-EM structures of thermostabilized prestin provide mechanistic insights underlying outer hair cell electromotility.
Nat Commun, 13, 2022
|
|
7V75
 
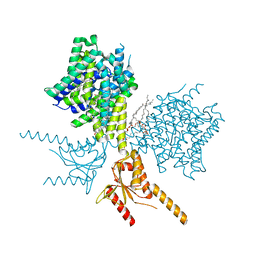 | | Thermostabilized human prestin in complex with salicylate | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 2-HYDROXYBENZOIC ACID, CHOLESTEROL, ... | | Authors: | Futamata, H, Fukuda, M, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-08-21 | | Release date: | 2022-08-31 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Cryo-EM structures of thermostabilized prestin provide mechanistic insights underlying outer hair cell electromotility.
Nat Commun, 13, 2022
|
|
7VKA
 
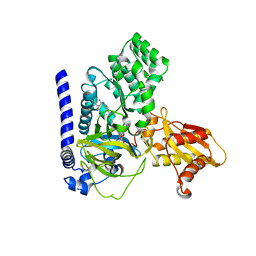 | | Crystal Structure of GH3.6 in complex with an inhibitor | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCEROL, Indole-3-acetic acid-amido synthetase GH3.6, ... | | Authors: | Wang, N, Luo, M, Bao, H, Huang, H. | | Deposit date: | 2021-09-29 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Chemical genetic screening identifies nalacin as an inhibitor of GH3 amido synthetase for auxin conjugation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7W8Y
 
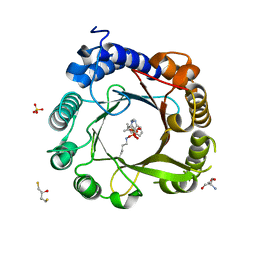 | | DMSPP- and Naplha-Me-Trp-bound 6-dimethylallyl tryptophan synthase, IptA | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-dimethylallyltryptophan synthase, ... | | Authors: | Suemune, H, Nagano, S. | | Deposit date: | 2021-12-08 | | Release date: | 2022-10-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Crystal structures of a 6-dimethylallyltryptophan synthase, IptA: Insights into substrate tolerance and enhancement of prenyltransferase activity.
Biochem.Biophys.Res.Commun., 593, 2022
|
|
7W8V
 
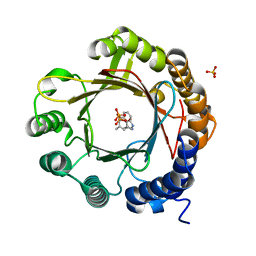 | | DMSPP- and Trp-bound 6-dimethylallyl tryptophan synthase, IptA | | Descriptor: | 6-dimethylallyltryptophan synthase, DIMETHYLALLYL S-THIOLODIPHOSPHATE, SULFATE ION, ... | | Authors: | Suemune, H, Nagano, S, Tomoya, H. | | Deposit date: | 2021-12-08 | | Release date: | 2022-10-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Crystal structures of a 6-dimethylallyltryptophan synthase, IptA: Insights into substrate tolerance and enhancement of prenyltransferase activity.
Biochem.Biophys.Res.Commun., 593, 2022
|
|
7W8X
 
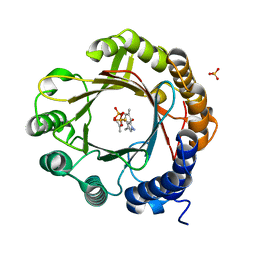 | | DMSPP- and 6-Me-Trp-bound dimethylallyl tryptophan synthase, IptA | | Descriptor: | (2S)-2-azanyl-3-(6-methyl-1H-indol-3-yl)propanoic acid, 6-dimethylallyltryptophan synthase, DIMETHYLALLYL S-THIOLODIPHOSPHATE, ... | | Authors: | Suemune, H, Nagano, S, Tomoya, H. | | Deposit date: | 2021-12-08 | | Release date: | 2022-10-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structures of a 6-dimethylallyltryptophan synthase, IptA: Insights into substrate tolerance and enhancement of prenyltransferase activity.
Biochem.Biophys.Res.Commun., 593, 2022
|
|
7W8W
 
 | | DMSPP- and 5-Me-Trp-bound 6-dimethylallyl tryptophan synthase, IptA | | Descriptor: | 5-methyl-L-tryptophan, 6-dimethylallyltryptophan synthase, DIMETHYLALLYL S-THIOLODIPHOSPHATE, ... | | Authors: | Suemune, H, Nagano, S, Tomoya, H. | | Deposit date: | 2021-12-08 | | Release date: | 2022-10-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of a 6-dimethylallyltryptophan synthase, IptA: Insights into substrate tolerance and enhancement of prenyltransferase activity.
Biochem.Biophys.Res.Commun., 593, 2022
|
|
7W8U
 
 | | Crystal Structure of Indole Prenyltransferase IptA | | Descriptor: | 6-dimethylallyltryptophan synthase | | Authors: | Suemune, H, Nagano, S. | | Deposit date: | 2021-12-08 | | Release date: | 2022-10-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structures of a 6-dimethylallyltryptophan synthase, IptA: Insights into substrate tolerance and enhancement of prenyltransferase activity.
Biochem.Biophys.Res.Commun., 593, 2022
|
|
