1KHE
 
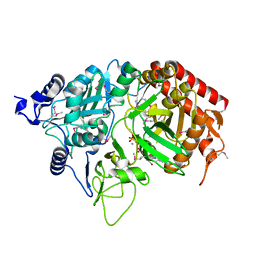 | | PEPCK complex with nonhydrolyzable GTP analog, MAD data | | Descriptor: | MANGANESE (II) ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, Phosphoenolpyruvate Carboxykinase, ... | | Authors: | Dunten, P, Belunis, C, Crowther, R, Hollfelder, K, Kammlott, U, Levin, W, Michel, H, Ramsey, G.B, Swain, A, Weber, D, Wertheimer, S.J. | | Deposit date: | 2001-11-29 | | Release date: | 2002-02-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of human cytosolic phosphoenolpyruvate carboxykinase reveals a new GTP-binding site.
J.Mol.Biol., 316, 2002
|
|
1KE9
 
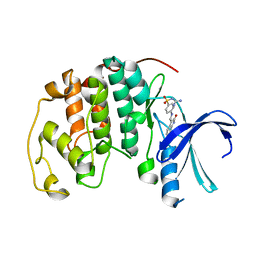 | | CYCLIN-DEPENDENT KINASE 2 (CDK2) COMPLEXED WITH 3-{[4-({[AMINO(IMINO)METHYL]AMINOSULFONYL)ANILINO]METHYLENE}-2-OXO-2,3-DIHYDRO-1H-INDOLE | | Descriptor: | 3-{[4-([AMINO(IMINO)METHYL]AMINOSULFONYL)ANILINO]METHYLENE}-2-OXO-2,3-DIHYDRO-1H-INDOLE, Cell division protein kinase 2 | | Authors: | Bramson, H.N, Corona, J, Davis, S.T, Dickerson, S.H, Edelstein, M, Frye, S.V, Gampe, R.T, Hassell, A.M, Shewchuk, L.M, Kuyper, L.F. | | Deposit date: | 2001-11-14 | | Release date: | 2002-05-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Oxindole-based inhibitors of cyclin-dependent kinase 2 (CDK2): design, synthesis, enzymatic activities, and X-ray crystallographic analysis.
J.Med.Chem., 44, 2001
|
|
1KHB
 
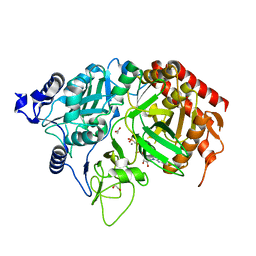 | | PEPCK complex with nonhydrolyzable GTP analog, native data | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, MANGANESE (II) ION, ... | | Authors: | Dunten, P, Belunis, C, Crowther, R, Hollfelder, K, Kammlott, U, Levin, W, Michel, H, Ramsey, G.B, Swain, A, Weber, D, Wertheimer, S.J. | | Deposit date: | 2001-11-29 | | Release date: | 2002-02-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.854 Å) | | Cite: | Crystal structure of human cytosolic phosphoenolpyruvate carboxykinase reveals a new GTP-binding site.
J.Mol.Biol., 316, 2002
|
|
1KE7
 
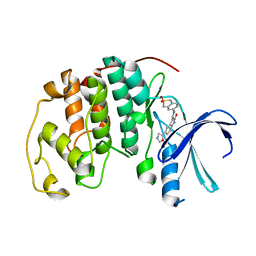 | | CYCLIN-DEPENDENT KINASE 2 (CDK2) COMPLEXED WITH 3-{[(2,2-DIOXIDO-1,3-DIHYDRO-2-BENZOTHIEN-5-YL)AMINO]METHYLENE}-5-(1,3-OXAZOL-5-YL)-1,3-DIHYDRO-2H-INDOL-2-ONE | | Descriptor: | 3-{[(2,2-DIOXIDO-1,3-DIHYDRO-2-BENZOTHIEN-5-YL)AMINO]METHYLENE}-5-(1,3-OXAZOL-5-YL)-1,3-DIHYDRO-2H-INDOL-2-ONE, Cell division protein kinase 2 | | Authors: | Bramson, H.N, Corona, J, Davis, S.T, Dickerson, S.H, Edelstein, M, Frye, S.V, Gampe, R.T, Hassell, A.H, Shewchuk, L.M, Kuyper, L.F. | | Deposit date: | 2001-11-14 | | Release date: | 2002-05-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Oxindole-based inhibitors of cyclin-dependent kinase 2 (CDK2): design, synthesis, enzymatic activities, and X-ray crystallographic analysis.
J.Med.Chem., 44, 2001
|
|
1KE5
 
 | | CDK2 complexed with N-methyl-4-{[(2-oxo-1,2-dihydro-3H-indol-3-ylidene)methyl]amino}benzenesulfonamide | | Descriptor: | Cell division protein kinase 2, N-METHYL-4-{[(2-OXO-1,2-DIHYDRO-3H-INDOL-3-YLIDENE)METHYL]AMINO}BENZENESULFONAMIDE | | Authors: | Bramson, H.N, Corona, J, Davis, S.T, Dickerson, S.H, Edelstein, M, Frye, S.V, Gampe, R.T, Hassell, A.M, Shewchuk, L.M, Kuyper, L.F. | | Deposit date: | 2001-11-14 | | Release date: | 2002-05-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Oxindole-based inhibitors of cyclin-dependent kinase 2 (CDK2): design, synthesis, enzymatic activities, and X-ray crystallographic analysis.
J.Med.Chem., 44, 2001
|
|
1KHG
 
 | | PEPCK | | Descriptor: | 1,2-ETHANEDIOL, MANGANESE (II) ION, Phosphoenolpyruvate carboxykinase, ... | | Authors: | Dunten, P, Belunis, C, Crowther, R, Hollfelder, K, Kammlott, U, Levin, W, Michel, H, Ramsey, G.B, Swain, A, Weber, D, Wertheimer, S.J. | | Deposit date: | 2001-11-29 | | Release date: | 2002-02-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Crystal structure of human cytosolic phosphoenolpyruvate carboxykinase reveals a new GTP-binding site.
J.Mol.Biol., 316, 2002
|
|
2DOQ
 
 | | crystal structure of Sfi1p/Cdc31p complex | | Descriptor: | CALCIUM ION, Cell division control protein 31, SFI1p | | Authors: | Li, S, Sandercock, A.M, Conduit, P.T, Robinson, C.V, Williams, R.L, Kilmartin, J.V. | | Deposit date: | 2006-05-03 | | Release date: | 2006-06-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural role of Sfi1p-centrin filaments in budding yeast spindle pole body duplication.
J.Cell Biol., 173, 2006
|
|
7M2U
 
 | | Nucleotide Excision Repair complex TFIIH Rad4-33 | | Descriptor: | CALCIUM ION, DNA repair helicase RAD25, DNA repair helicase RAD3, ... | | Authors: | van Eeuwen, T, Murakami, K. | | Deposit date: | 2021-03-17 | | Release date: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | Cryo-EM structure of TFIIH/Rad4-Rad23-Rad33 in damaged DNA opening in nucleotide excision repair.
Nat Commun, 12, 2021
|
|
6V92
 
 | | RSC-NCP | | Descriptor: | Actin-like protein ARP9, Actin-related protein 7, Chromatin structure-remodeling complex protein RSC14, ... | | Authors: | Patel, A.B, Moore, C.M, Greber, B.J, Nogales, E. | | Deposit date: | 2019-12-13 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Architecture of the chromatin remodeler RSC and insights into its nucleosome engagement.
Elife, 8, 2019
|
|
6K1P
 
 | | The complex of ISWI-nucleosome in the ADP.BeF-bound state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA (167-MER), ... | | Authors: | Yan, L.J, Wu, H, Li, X.M, Gao, N, Chen, Z.C. | | Deposit date: | 2019-05-10 | | Release date: | 2019-05-29 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | Structures of the ISWI-nucleosome complex reveal a conserved mechanism of chromatin remodeling.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6JYL
 
 | | The crosslinked complex of ISWI-nucleosome in the ADP.BeF-bound state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA (167-MER), ... | | Authors: | Yan, L.J, Wu, H, Li, X.M, Gao, N, Chen, Z.C. | | Deposit date: | 2019-04-26 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Structures of the ISWI-nucleosome complex reveal a conserved mechanism of chromatin remodeling.
Nat.Struct.Mol.Biol., 26, 2019
|
|
9BGM
 
 | | Pseudomonas phage DEV neck and tail (portal, head-to-tail and tail tube proteins) | | Descriptor: | gp75 tail tube, gp80 portal protein, gp83 head-to-tail | | Authors: | Iglesias, S.M, Hou, C.-F.D, Li, F, Cingolani, G. | | Deposit date: | 2024-04-19 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Integrative structural analysis of Pseudomonas phage DEV reveals a genome ejection motor.
Nat Commun, 15, 2024
|
|
9COD
 
 | | C15 symmetrized DEV collar | | Descriptor: | SGNH hydrolase-type esterase domain-containing protein | | Authors: | Iglesias, S.M, Hou, C.F.D, Li, F, Cingolani, G. | | Deposit date: | 2024-07-16 | | Release date: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Integrative structural analysis of Pseudomonas phage DEV reveals a genome ejection motor.
Nat Commun, 15, 2024
|
|
9BGO
 
 | |
9BGN
 
 | |
4UXN
 
 | | LSD1(KDM1A)-CoREST in complex with Z-Pro derivative of MC2580 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, REST COREPRESSOR 1, ... | | Authors: | Rodriguez, V, Valente, S, Stazi, G, Lucidi, A, Mercurio, C, Vianello, P, Ciossani, G, Mattevi, A, Botrugno, O.A, Dessanti, P, Minucci, S, Varasi, M, Mai, A. | | Deposit date: | 2014-08-27 | | Release date: | 2015-02-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Pyrrole- and Indole-Containing Tranylcypromine Derivatives as Novel Lysine-Specific Demethylase 1 Inhibitors Active on Cancer Cells
Chemmedchem, 6, 2015
|
|
7SSA
 
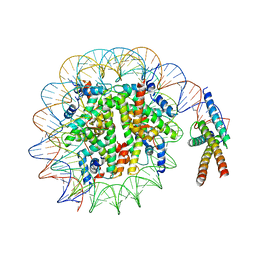 | | Cryo-EM structure of pioneer factor Cbf1 bound to the nucleosome | | Descriptor: | Centromere-binding protein 1, DNA (137-MER), Histone H2A.1, ... | | Authors: | Eek, P, Tan, S. | | Deposit date: | 2021-11-10 | | Release date: | 2023-04-05 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Basic helix-loop-helix pioneer factors interact with the histone octamer to invade nucleosomes and generate nucleosome-depleted regions.
Mol.Cell, 83, 2023
|
|
7P15
 
 | |
7OZW
 
 | |
7AO9
 
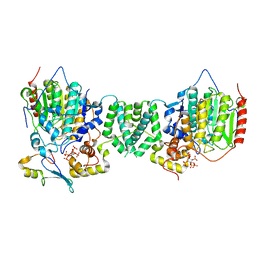 | | Structure of the core MTA1/HDAC1/MBD2 NURD deacetylase complex | | Descriptor: | Histone deacetylase 1, INOSITOL HEXAKISPHOSPHATE, Metastasis-associated protein MTA1, ... | | Authors: | Millard, C.J, Fairall, L, Ragan, T.J, Savva, C.G, Schwabe, J.W.R. | | Deposit date: | 2020-10-14 | | Release date: | 2020-11-11 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | The topology of chromatin-binding domains in the NuRD deacetylase complex.
Nucleic Acids Res., 48, 2020
|
|
7AO8
 
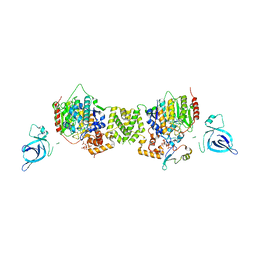 | | Structure of the MTA1/HDAC1/MBD2 NURD deacetylase complex | | Descriptor: | Histone deacetylase 1, INOSITOL HEXAKISPHOSPHATE, Metastasis-associated protein MTA1, ... | | Authors: | Millard, C.J, Fairall, L, Ragan, T.J, Savva, C.G, Schwabe, J.W.R. | | Deposit date: | 2020-10-14 | | Release date: | 2020-11-11 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | The topology of chromatin-binding domains in the NuRD deacetylase complex.
Nucleic Acids Res., 48, 2020
|
|
8X19
 
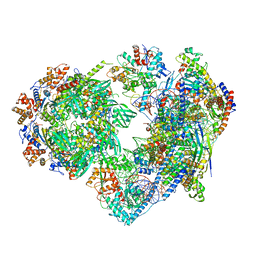 | | Structure of nucleosome-bound SRCAP-C in the ADP-BeFx-bound state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Yu, J, Wang, Q, Yu, Z, Li, W, Wang, L, Xu, Y. | | Deposit date: | 2023-11-06 | | Release date: | 2024-03-06 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into histone exchange by human SRCAP complex.
Cell Discov, 10, 2024
|
|
8X15
 
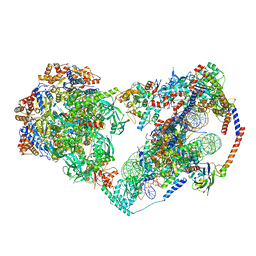 | | Structure of nucleosome-bound SRCAP-C in the apo state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Yu, J, Wang, Q, Yu, Z, Li, W, Wang, L, Xu, Y. | | Deposit date: | 2023-11-06 | | Release date: | 2024-03-06 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into histone exchange by human SRCAP complex.
Cell Discov, 10, 2024
|
|
8X1C
 
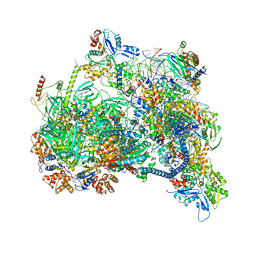 | | Structure of nucleosome-bound SRCAP-C in the ADP-bound state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Yu, J, Wang, Q, Yu, Z, Li, W, Wang, L, Xu, Y. | | Deposit date: | 2023-11-06 | | Release date: | 2024-03-06 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into histone exchange by human SRCAP complex.
Cell Discov, 10, 2024
|
|
8B3I
 
 | | CRL4CSA-E2-Ub (state 2) | | Descriptor: | Cullin-4A, DNA damage-binding protein 1, DNA excision repair protein ERCC-8, ... | | Authors: | Kokic, G, Cramer, P. | | Deposit date: | 2022-09-16 | | Release date: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | C(N)RL4CSA-E2-Ub (state 2)
To Be Published
|
|
