7YL0
 
 | | Structure of hIAPP-TF-type2 | | Descriptor: | Islet amyloid polypeptide | | Authors: | Li, D, Zhang, X, Wang, Y, Zhu, P. | | Deposit date: | 2022-07-25 | | Release date: | 2022-12-28 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A new polymorphism of human amylin fibrils with similar protofilaments and a conserved core.
Iscience, 25, 2022
|
|
7YKW
 
 | |
7YKV
 
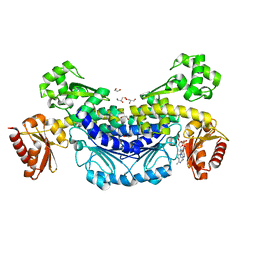 | | The structure of archaeal nuclease RecJ2 from Methanocaldococcus jannaschii | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, [5-(6-aminopurin-9-yl)-2-(hydroxymethyl)furan-3-yl] [5-(6-aminopurin-9-yl)-3-oxidanyl-furan-2-yl]methyl hydrogen phosphate, ... | | Authors: | Wang, W.W, Liu, X.P. | | Deposit date: | 2022-07-24 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | The structure of archaeal nuclease RecJ2 from Methanocaldococcus jannaschii
To Be Published
|
|
7YKP
 
 | | Crystal structure of a novel alpha/beta hydrolase from thermomonospora curvata with glycerol | | Descriptor: | GLYCEROL, Triacylglycerol lipase | | Authors: | Han, X, Jian, G, Bornscheuer, U.T, Wei, R, Liu, W. | | Deposit date: | 2022-07-23 | | Release date: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Crystal structure of a novel alpha/beta hydrolase from thermomonospora curvata with glycerol
To Be Published
|
|
7YKN
 
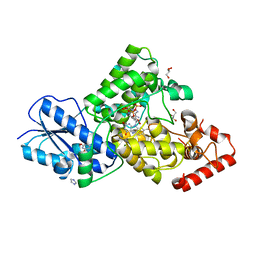 | | Crystal structure of (6-4) photolyase from Vibrio cholerae | | Descriptor: | 1-deoxy-1-(6,7-dimethyl-2,4-dioxo-3,4-dihydropteridin-8(2H)-yl)-D-ribitol, Cryptochrome/photolyase family protein, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Cakilkaya, B, Kavakli, I.H, DeMirci, H. | | Deposit date: | 2022-07-23 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of Vibrio cholerae (6-4) photolyase reveals interactions with cofactors and a DNA-binding region.
J.Biol.Chem., 299, 2023
|
|
7YKI
 
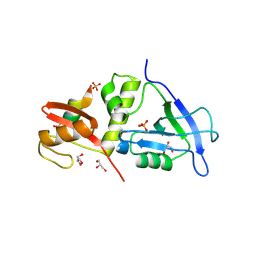 | | Crystal structure of MAGI2 PDZ0-GK domain in complex with phospho-SAPAP1 GBR3 peptide | | Descriptor: | GLYCEROL, Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 2, ... | | Authors: | Zhang, M, Lin, L, Zhu, J. | | Deposit date: | 2022-07-22 | | Release date: | 2023-08-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Phosphorylation-dependent recognition of diverse protein targets by the cryptic GK domain of MAGI MAGUKs.
Sci Adv, 9, 2023
|
|
7YKH
 
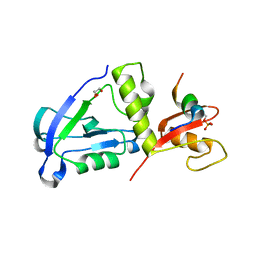 | | Crystal structure of MAGI2 PDZ0-GK domain in complex with phospho-SAPAP1 GBR2 peptide | | Descriptor: | GLYCEROL, Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 2, ... | | Authors: | Zhang, M, Lin, L, Zhu, J. | | Deposit date: | 2022-07-22 | | Release date: | 2023-08-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Phosphorylation-dependent recognition of diverse protein targets by the cryptic GK domain of MAGI MAGUKs.
Sci Adv, 9, 2023
|
|
7YKB
 
 | | Neutron Structure of PcyA D105N Mutant Complexed with Biliverdin at Room Temperature | | Descriptor: | 3-[5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-2-[[5-[(Z)-(3-ethenyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1H-pyrrol-2-yl]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, Phycocyanobilin:ferredoxin oxidoreductase, SODIUM ION | | Authors: | Unno, M, Nanasawa, R. | | Deposit date: | 2022-07-22 | | Release date: | 2023-01-25 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (1.38 Å), X-RAY DIFFRACTION | | Cite: | Neutron crystallography and quantum chemical analysis of bilin reductase PcyA mutants reveal substrate and catalytic residue protonation states.
J.Biol.Chem., 299, 2022
|
|
7YK9
 
 | | Neutron Structure of PcyA I86D Mutant Complexed with Biliverdin at Room Temperature | | Descriptor: | 3-[5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-2-[[5-[(Z)-(3-ethenyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1H-pyrrol-2-yl]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, Phycocyanobilin:ferredoxin oxidoreductase | | Authors: | Unno, M, Igarashi, K. | | Deposit date: | 2022-07-22 | | Release date: | 2023-01-25 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (1.9 Å), X-RAY DIFFRACTION | | Cite: | Neutron crystallography and quantum chemical analysis of bilin reductase PcyA mutants reveal substrate and catalytic residue protonation states.
J.Biol.Chem., 299, 2022
|
|
7YK7
 
 | | Cryo-EM structure of the DC591053-bound human relaxin family peptide receptor 4 (RXFP4)-Gi complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-2, ... | | Authors: | Chen, Y, Zhou, Q.T, Wang, J, Xu, Y.W, Wang, Y, Yan, J.H, Wang, Y.B, Zhu, Q, Zhao, F.H, Li, C.H, Chen, C.W, Cai, X.Q, Bathgate, R.A.D, Shen, C, Xu, H.E, Yang, D.H, Liu, H, Wang, M.W. | | Deposit date: | 2022-07-21 | | Release date: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Ligand recognition mechanism of the human relaxin family peptide receptor 4 (RXFP4).
Nat Commun, 14, 2023
|
|
7YK6
 
 | | Cryo-EM structure of the compound 4-bound human relaxin family peptide receptor 4 (RXFP4)-Gi complex | | Descriptor: | 1-[2-(4-chlorophenyl)ethyl]-3-[(7-ethyl-5-oxidanyl-1H-indol-3-yl)methylideneamino]guanidine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Chen, Y, Zhou, Q.T, Wang, J, Xu, Y.W, Wang, Y, Yan, J.H, Wang, Y.B, Zhu, Q, Zhao, F.H, Li, C.H, Chen, C.W, Cai, X.Q, Bathgate, R.A.D, Shen, C, Liu, H, Xu, H.E, Yang, D.H, Wang, M.W. | | Deposit date: | 2022-07-21 | | Release date: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Ligand recognition mechanism of the human relaxin family peptide receptor 4 (RXFP4).
Nat Commun, 14, 2023
|
|
7YJL
 
 | |
7YJH
 
 | | Crystal structure of the P450 BM3 heme domain mutant F87V/T268I in complex with N-imidazolyl-pentanoyl-L-phenylalanine and hydroxylamine | | Descriptor: | (2~{S})-2-(5-imidazol-1-ylpentanoylamino)-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, HYDROXYAMINE, ... | | Authors: | Dong, S, Chen, J, Jiang, Y, Cong, Z, Feng, Y. | | Deposit date: | 2022-07-20 | | Release date: | 2023-05-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | Regiodivergent and Enantioselective Hydroxylation of C-H bonds by Synergistic Use of Protein Engineering and Exogenous Dual-Functional Small Molecules.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7YJG
 
 | | Crystal structure of the P450 BM3 heme domain mutant F87A/T268V/A82C/L181M in complex with N-imidazolyl-pentanoyl-L-phenylalanine and hydroxylamine | | Descriptor: | (2~{S})-2-(5-imidazol-1-ylpentanoylamino)-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, HYDROXYAMINE, ... | | Authors: | Dong, S, Chen, J, Jiang, Y, Cong, Z, Feng, Y. | | Deposit date: | 2022-07-20 | | Release date: | 2023-05-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.684 Å) | | Cite: | Regiodivergent and Enantioselective Hydroxylation of C-H bonds by Synergistic Use of Protein Engineering and Exogenous Dual-Functional Small Molecules.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7YJF
 
 | | Crystal structure of the P450 BM3 heme domain mutant F87A/T268P/V78I in complex with N-imidazolyl-pentanoyl-L-phenylalanine and hydroxylamine | | Descriptor: | (2~{S})-2-(5-imidazol-1-ylpentanoylamino)-3-phenyl-propanoic acid, HYDROXYAMINE, P450 BM3 heme domain mutant F87A/T268P/V78I, ... | | Authors: | Dong, S, Chen, J, Jiang, Y, Cong, Z, Feng, Y. | | Deposit date: | 2022-07-20 | | Release date: | 2023-05-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.515 Å) | | Cite: | Regiodivergent and Enantioselective Hydroxylation of C-H bonds by Synergistic Use of Protein Engineering and Exogenous Dual-Functional Small Molecules.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7YJE
 
 | | Crystal structure of the P450 BM3 heme domain mutant F87G/T268V/A184V/A328V in complex with N-imidazolyl-hexanoyl-L-phenylalanine and acetate ion | | Descriptor: | (2S)-2-(6-imidazol-1-ylhexanoylamino)-3-phenyl-propanoic acid, ACETATE ION, Bifunctional cytochrome P450/NADPH--P450 reductase, ... | | Authors: | Dong, S, Chen, J, Jiang, Y, Cong, Z, Feng, Y. | | Deposit date: | 2022-07-20 | | Release date: | 2023-05-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Regiodivergent and Enantioselective Hydroxylation of C-H bonds by Synergistic Use of Protein Engineering and Exogenous Dual-Functional Small Molecules.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7YJD
 
 | | Crystal structure of the P450 BM3 heme domain mutant F87A in complex with N-imidazolyl-hexanoyl-L-phenylalanine and hydroxylamine | | Descriptor: | (2S)-2-(6-imidazol-1-ylhexanoylamino)-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, HYDROXYAMINE, ... | | Authors: | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | Deposit date: | 2022-07-19 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the P450 BM3 heme domain mutant F87A in complex with N-imidazolyl-hexanoyl-L-phenylalanine and hydroxylamine
To Be Published
|
|
7YJB
 
 | | Crystal structure of Stenoloma chusanum chalcone synthase 1 (ScCHS1) complex with CoA and Eriodictyol | | Descriptor: | (2S)-2-(3,4-DIHYDROXYPHENYL)-5,7-DIHYDROXY-2,3-DIHYDRO-4H-CHROMEN-4-ONE, COENZYME A, chalcone synthase 1 (ScCHS1) | | Authors: | Li, J.X, Cheng, A.X, Zhang, P. | | Deposit date: | 2022-07-19 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular and structural characterization of a promiscuous chalcone synthase from the fern species Stenoloma chusanum.
J Integr Plant Biol, 64, 2022
|
|
7YJA
 
 | |
7YJ9
 
 | |
7YJ8
 
 | |
7YJ7
 
 | |
7YJ6
 
 | |
7YJ5
 
 | |
7YJ4
 
 | | Cryo-EM structure of the INSL5-bound human relaxin family peptidereceptor 4 (RXFP4)-Gi complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-2, ... | | Authors: | Chen, Y, Zhou, Q.T, Wang, J, Xu, Y.W, Wang, Y, Yan, J.H, Wang, Y.B, Zhu, Q, Zhao, F.H, Li, C.H, Chen, C.W, Cai, X.Q, Bathgate, R.A.D, Shen, C, Liu, H, Xu, H.E, Yang, D.H, Wang, M.W. | | Deposit date: | 2022-07-19 | | Release date: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Ligand recognition mechanism of the human relaxin family peptide receptor 4 (RXFP4).
Nat Commun, 14, 2023
|
|
