6S0R
 
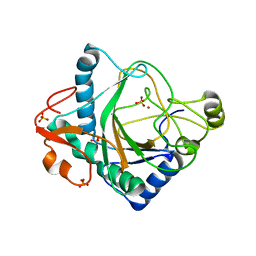 | | The crystal structure of kanamycin B dioxygenase (KanJ) from Streptomyces kanamyceticus complex with nickel, sulfate and chloride | | Descriptor: | CHLORIDE ION, Kanamycin B dioxygenase, NICKEL (II) ION, ... | | Authors: | Mrugala, B, Porebski, P.J, Niedzialkowska, E, Cymborowski, M.T, Minor, W, Borowski, T. | | Deposit date: | 2019-06-18 | | Release date: | 2020-07-08 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A study on the structure, mechanism, and biochemistry of kanamycin B dioxygenase (KanJ)-an enzyme with a broad range of substrates.
Febs J., 288, 2021
|
|
6M11
 
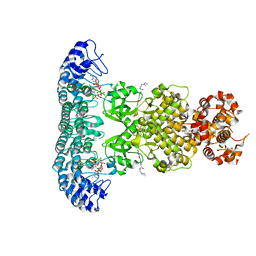 | | Crystal structure of Rnase L in complex with Sunitinib | | Descriptor: | 5'-O-MONOPHOSPHORYLADENYLYL(2'->5')ADENYLYL(2'->5')ADENOSINE, N-[2-(diethylamino)ethyl]-5-[(Z)-(5-fluoro-2-oxo-1,2-dihydro-3H-indol-3-ylidene)methyl]-2,4-dimethyl-1H-pyrrole-3-carbo xamide, PHOSPHATE ION, ... | | Authors: | Tang, J, Huang, H. | | Deposit date: | 2020-02-24 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Sunitinib inhibits RNase L by destabilizing its active dimer conformation.
Biochem.J., 477, 2020
|
|
6M12
 
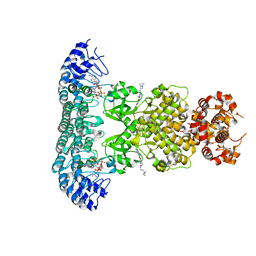 | | Crystal Structure of Rnase L in complex with SU11652 | | Descriptor: | 5-[(E)-(5-CHLORO-2-OXO-1,2-DIHYDRO-3H-INDOL-3-YLIDENE)METHYL]-N-[2-(DIETHYLAMINO)ETHYL]-2,4-DIMETHYL-1H-PYRROLE-3-CARBOXAMIDE, PHOSPHATE ION, Ribonuclease L, ... | | Authors: | Tang, J, Huang, H. | | Deposit date: | 2020-02-24 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Sunitinib inhibits RNase L by destabilizing its active dimer conformation.
Biochem.J., 477, 2020
|
|
6M13
 
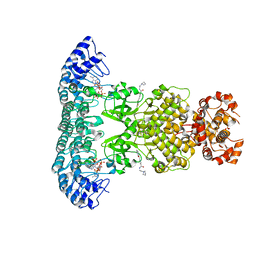 | | Crystal structure of Rnase L in complex with Toceranib | | Descriptor: | 5-[(Z)-(5-fluoranyl-2-oxidanylidene-1H-indol-3-ylidene)methyl]-2,4-dimethyl-N-(2-pyrrolidin-1-ylethyl)-1H-pyrrole-3-carboxamide, PHOSPHATE ION, Ribonuclease L, ... | | Authors: | Tang, J, Huang, H. | | Deposit date: | 2020-02-24 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Sunitinib inhibits RNase L by destabilizing its active dimer conformation.
Biochem.J., 477, 2020
|
|
9NXH
 
 | | An alpha-l-arabinofuranosidase (AtAbf43C) from Acetivibrio thermocellus DSM1313 bound to arabinofuranose | | Descriptor: | GLYCEROL, Glycoside hydrolase family 43, MAGNESIUM ION, ... | | Authors: | Galindo, J.L, Jeffrey, P.D, Conway, J.M. | | Deposit date: | 2025-03-25 | | Release date: | 2025-08-20 | | Last modified: | 2025-10-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Functional and structural characterization of AtAbf43C: an exo-1,5-alpha-L-arabinofuranosidase from Acetivibrio thermocellus DSM1313.
Biochem.J., 482, 2025
|
|
9MNN
 
 | | Crystal structure of L. monocytogenes MenD with Mg2+ and intermediate I bound | | Descriptor: | (4S)-4-{3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-5-(2-{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}ethyl)-4-methyl-1,3lambda~5~-thiazol-2-yl}-4-hydroxybutanoic acid, 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylate synthase, MAGNESIUM ION | | Authors: | Klein, M, Given, F.M, Ho, N.A.T, Allison, T.M, Johnston, J.M. | | Deposit date: | 2024-12-22 | | Release date: | 2025-07-30 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structures of Listeria monocytogenes MenD in ThDP-bound and in-crystallo captured intermediate I-bound forms.
Acta Crystallogr.,Sect.F, 81, 2025
|
|
8BWU
 
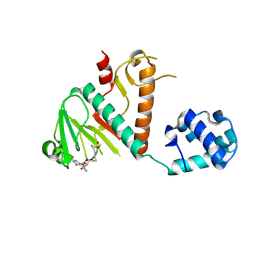 | | Crystal structure of SARS-CoV-2 nsp14 methyltransferase domain in complex with the SS148 inhibitor | | Descriptor: | (2~{S})-2-azanyl-4-[[(2~{S},3~{S},4~{R},5~{R})-5-(4-azanyl-5-cyano-pyrrolo[2,3-d]pyrimidin-7-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanyl]butanoic acid, Transcription factor ETV6,Proofreading exoribonuclease nsp14, ZINC ION | | Authors: | Konkolova, E, Klima, M, Boura, E, Jin, J, Kaniskan, H.U, Han, Y, Vedadi, M. | | Deposit date: | 2022-12-07 | | Release date: | 2023-10-11 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Application of established computational techniques to identify potential SARS-CoV-2 Nsp14-MTase inhibitors in low data regimes
Digit Discov, 2024
|
|
9NXJ
 
 | | The GH43 domain of an alpha-l-arabinofuranosidase (AtAbf43C_GH43) from Acetivibrio thermocellus DSM1313 | | Descriptor: | Glycoside hydrolase family 43, SULFATE ION, alpha-L-arabinofuranose | | Authors: | Galindo, J.L, Jeffrey, P.D, Conway, J.M. | | Deposit date: | 2025-03-25 | | Release date: | 2025-08-20 | | Last modified: | 2025-10-15 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Functional and structural characterization of AtAbf43C: an exo-1,5-alpha-L-arabinofuranosidase from Acetivibrio thermocellus DSM1313.
Biochem.J., 482, 2025
|
|
9CQE
 
 | |
2VXW
 
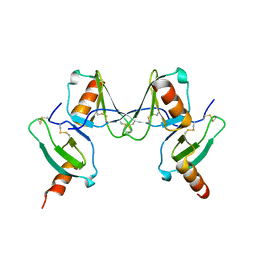 | |
6JWW
 
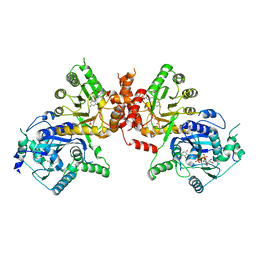 | | Crystal structure of Plasmodium falciparum HPPK-DHPS S436F/A437G/A613T triple mutant with STZ-DHP | | Descriptor: | 2-AMINO-6-HYDROXYMETHYL-7,8-DIHYDRO-3H-PTERIDIN-4-ONE, 4-{[(2-amino-4-oxo-3,4,7,8-tetrahydropteridin-6-yl)methyl]amino}-N-(1,3-thiazol-2-yl)benzenesulfonamide, 7,8-dihydro-6-hydroxymethylpterin pyrophosphokinase-dihydropteroate synthase, ... | | Authors: | Chitnumsub, P, Jaruwat, A, Yuthavong, Y. | | Deposit date: | 2019-04-21 | | Release date: | 2020-02-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The structure of Plasmodium falciparum hydroxymethyldihydropterin pyrophosphokinase-dihydropteroate synthase reveals the basis of sulfa resistance.
Febs J., 287, 2020
|
|
5LUD
 
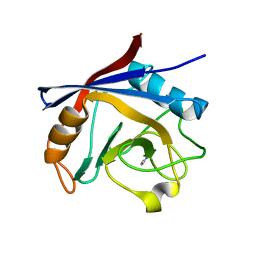 | | Structure of Cyclophilin A in complex with 2,3-Diaminopyridine | | Descriptor: | Peptidyl-prolyl cis-trans isomerase, pyridine-2,3-diamine | | Authors: | McNae, I.W, Nowicki, M.W, Blackburn, E.A, Wear, M.A, Walkinshaw, M.D. | | Deposit date: | 2016-09-08 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Thermo-kinetic analysis space expansion for cyclophilin-ligand interactions - identification of a new nonpeptide inhibitor using BiacoreTM T200.
FEBS Open Bio, 7, 2017
|
|
6JWY
 
 | | Crystal structure of Plasmodium falciparum HPPK-DHPS A437G with SDX-DHP | | Descriptor: | 2-AMINO-6-HYDROXYMETHYL-7,8-DIHYDRO-3H-PTERIDIN-4-ONE, 4-[(2-azanyl-4-oxidanylidene-7,8-dihydro-3~{H}-pteridin-6-yl)methylamino]-~{N}-(5,6-dimethoxypyrimidin-4-yl)benzenesulfonamide, 7,8-dihydro-6-hydroxymethylpterin pyrophosphokinase-dihydropteroate synthase, ... | | Authors: | Chitnumsub, P, Jaruwat, A, Yuthavong, Y. | | Deposit date: | 2019-04-21 | | Release date: | 2020-02-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of Plasmodium falciparum hydroxymethyldihydropterin pyrophosphokinase-dihydropteroate synthase reveals the basis of sulfa resistance.
Febs J., 287, 2020
|
|
6JWX
 
 | | Crystal structure of Plasmodium falciparum HPPK-DHPS wild type with SDX-DHP | | Descriptor: | 2-AMINO-6-HYDROXYMETHYL-7,8-DIHYDRO-3H-PTERIDIN-4-ONE, 4-[(2-azanyl-4-oxidanylidene-7,8-dihydro-3~{H}-pteridin-6-yl)methylamino]-~{N}-(5,6-dimethoxypyrimidin-4-yl)benzenesulfonamide, 7,8-dihydro-6-hydroxymethylpterin pyrophosphokinase-dihydropteroate synthase, ... | | Authors: | Chitnumsub, P, Jaruwat, A, Yuthavong, Y. | | Deposit date: | 2019-04-21 | | Release date: | 2020-02-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of Plasmodium falciparum hydroxymethyldihydropterin pyrophosphokinase-dihydropteroate synthase reveals the basis of sulfa resistance.
Febs J., 287, 2020
|
|
6Q0N
 
 | | Structure of the Erbin PDB domain in complex with a high-affinity peptide | | Descriptor: | Erbin, peptide | | Authors: | Singer, A.U, Teyra, J, Ernst, A, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2019-08-02 | | Release date: | 2019-11-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Comprehensive analysis of all evolutionary paths between two divergent PDZ domain specificities.
Protein Sci., 29, 2020
|
|
6SV3
 
 | | Structure of coproheme-LmCpfC | | Descriptor: | 1,3,5,8-TETRAMETHYL-PORPHINE-2,4,6,7-TETRAPROPIONIC ACID FERROUS COMPLEX, Ferrochelatase, GLYCEROL | | Authors: | Hofbauer, S, Helm, J, Djinovic-Carugo, K, Furtmueller, P.G. | | Deposit date: | 2019-09-17 | | Release date: | 2019-12-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.64000869 Å) | | Cite: | Crystal structures and calorimetry reveal catalytically relevant binding mode of coproporphyrin and coproheme in coproporphyrin ferrochelatase.
Febs J., 287, 2020
|
|
6KFZ
 
 | | SufS from Bacillus subtilis, soaked with L-cysteine for 90 sec at 1.96 angstrom resolution | | Descriptor: | Cysteine desulfurase SufS, DI(HYDROXYETHYL)ETHER, N-({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)-L-CYSTEINE | | Authors: | Nakamura, R, Takahashi, Y, Fujishiro, T. | | Deposit date: | 2019-07-09 | | Release date: | 2019-10-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Snapshots of PLP-substrate and PLP-product external aldimines as intermediates in two types of cysteine desulfurase enzymes.
Febs J., 287, 2020
|
|
6JWS
 
 | | Crystal structure of Plasmodium falciparum HPPK-DHPS A437G with Pteroate | | Descriptor: | 7,8-dihydro-6-hydroxymethylpterin pyrophosphokinase-dihydropteroate synthase, ACETATE ION, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Chitnumsub, P, Jaruwat, A, Yuthavong, Y. | | Deposit date: | 2019-04-21 | | Release date: | 2020-02-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of Plasmodium falciparum hydroxymethyldihydropterin pyrophosphokinase-dihydropteroate synthase reveals the basis of sulfa resistance.
Febs J., 287, 2020
|
|
6KLK
 
 | |
6KUC
 
 | | Crystal structure of Plasmodium falciparum histo-aspartic protease (HAP) zymogen (Form 2) | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, HAP protein | | Authors: | Rathore, I, Mishra, V, Bhaumik, P. | | Deposit date: | 2019-08-31 | | Release date: | 2020-05-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Activation mechanism of plasmepsins, pepsin-like aspartic proteases from Plasmodium, follows a unique trans-activation pathway.
Febs J., 288, 2021
|
|
2X06
 
 | | SULFOLACTATE DEHYDROGENASE FROM METHANOCALDOCOCCUS JANNASCHII | | Descriptor: | L-SULFOLACTATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Irimia, A, Madern, D, Zaccai, G, Vellieux, F.M.D. | | Deposit date: | 2009-12-07 | | Release date: | 2009-12-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Methanoarchaeal Sulfolactate Dehydrogenase: Prototype of a New Family of Nadh-Dependent Enzymes.
Embo J., 23, 2004
|
|
6KUD
 
 | | Crystal structure of Plasmodium falciparum histo-aspartic protease (HAP) zymogen (Form 3) | | Descriptor: | GLYCEROL, HAP protein | | Authors: | Rathore, I, Mishra, V, Bhaumik, P. | | Deposit date: | 2019-08-31 | | Release date: | 2020-05-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Activation mechanism of plasmepsins, pepsin-like aspartic proteases from Plasmodium, follows a unique trans-activation pathway.
Febs J., 288, 2021
|
|
6KUB
 
 | | Crystal structure of Plasmodium falciparum histo-aspartic protease (HAP) zymogen (Form 1) | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, HAP protein | | Authors: | Rathore, I, Mishra, V, Bhaumik, P. | | Deposit date: | 2019-08-31 | | Release date: | 2020-05-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Activation mechanism of plasmepsins, pepsin-like aspartic proteases from Plasmodium, follows a unique trans-activation pathway.
Febs J., 288, 2021
|
|
6L0I
 
 | | Crystal structure of dihydroorotase in complex with malate at pH6.5 from Saccharomyces cerevisiae | | Descriptor: | (2S)-2-hydroxybutanedioic acid, Dihydroorotase, ZINC ION | | Authors: | Guan, H.H, Huang, Y.H, Huang, C.Y, Chen, C.J. | | Deposit date: | 2019-09-26 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the interaction modes of dihydroorotase with the anticancer drugs 5-fluorouracil and 5-aminouracil.
Biochem.Biophys.Res.Commun., 551, 2021
|
|
5MQQ
 
 | |
