8SDH
 
 | |
4WJT
 
 | | Stationary Phase Survival Protein YuiC from B.subtilis complexed with NAG | | Descriptor: | (2S)-2-{[(2S)-2-{[(2R)-2-hydroxypropyl]oxy}propyl]oxy}propan-1-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Quay, D.H.X, Cole, A.R, Cryar, A, Thalassinos, K, Williams, M.A, Bhakta, S, Keep, N.H. | | Deposit date: | 2014-10-01 | | Release date: | 2015-07-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Structure of the stationary phase survival protein YuiC from B.subtilis.
Bmc Struct.Biol., 15, 2015
|
|
6DQ4
 
 | | LINKED KDM5A JMJ DOMAIN BOUND TO THE INHIBITOR GSK-J1 | | Descriptor: | 3-[[2-pyridin-2-yl-6-(1,2,4,5-tetrahydro-3-benzazepin-3-yl)pyrimidin-4-yl]amino]propanoic acid, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2018-06-10 | | Release date: | 2018-11-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.392 Å) | | Cite: | Structure-Based Engineering of Irreversible Inhibitors against Histone Lysine Demethylase KDM5A.
J. Med. Chem., 61, 2018
|
|
6DFS
 
 | | mouse TCR I.29 in complex with IAg7-p8E9E6ss | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, H-2 class II histocompatibility antigen, A-D alpha chain, ... | | Authors: | Wang, Y, Dai, S. | | Deposit date: | 2018-05-15 | | Release date: | 2019-04-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | How C-terminal additions to insulin B-chain fragments create superagonists for T cells in mouse and human type 1 diabetes.
Sci Immunol, 4, 2019
|
|
4WLN
 
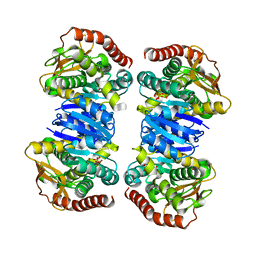 | | Crystal structure of apo MDH2 | | Descriptor: | Malate dehydrogenase, mitochondrial, PHOSPHATE ION | | Authors: | Eo, Y.M, Han, B.G, Ahn, H.C. | | Deposit date: | 2014-10-07 | | Release date: | 2015-11-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structure of apo MDH2
To Be Published
|
|
4WJG
 
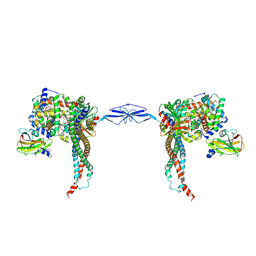 | | Structure of T. brucei haptoglobin-hemoglobin receptor binding to human haptoglobin-hemoglobin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Haptoglobin, ... | | Authors: | Stoedkilde, K, Torvund-Jensen, M, Moestrup, S.K, Andersen, C.B.F. | | Deposit date: | 2014-09-30 | | Release date: | 2014-11-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for trypanosomal haem acquisition and susceptibility to the host innate immune system.
Nat Commun, 5, 2014
|
|
8THM
 
 | |
8TFP
 
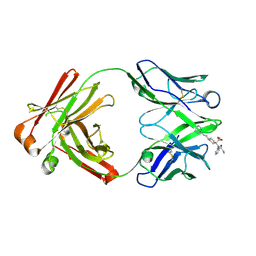 | | Fab from C10-S66K antibody in complex with carfentanil | | Descriptor: | Carfentanil, Heavy chain of Fab from C10-S66K antibody, Light chain of Fab from C10-S66K antibody | | Authors: | Pholcharee, T, Wilson, I.A. | | Deposit date: | 2023-07-11 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | An Engineered Human-Antibody Fragment with Fentanyl Pan-Specificity That Reverses Carfentanil-Induced Respiratory Depression.
Acs Chem Neurosci, 14, 2023
|
|
8TGD
 
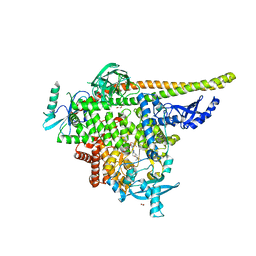 | | STX-478, a Mutant-Selective, Allosteric Inhibitor bound to H1047R PI3Kalpha | | Descriptor: | 1,2-ETHANEDIOL, N-(2-aminopyrimidin-5-yl)-N'-[(1R)-1-(5,7-difluoro-3-methyl-1-benzofuran-2-yl)-2,2,2-trifluoroethyl]urea, N~2~-{(4S,11aP)-2-[(4S)-4-(difluoromethyl)-2-oxo-1,3-oxazolidin-3-yl]-5,6-dihydroimidazo[1,2-d][1,4]benzoxazepin-9-yl}-L-alaninamide, ... | | Authors: | Hilbert, B, Brooijmans, N, Buckbinder, L, St.Jean Jr, D.J. | | Deposit date: | 2023-07-12 | | Release date: | 2023-09-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.928 Å) | | Cite: | STX-478, a Mutant-Selective, Allosteric PI3K alpha Inhibitor Spares Metabolic Dysfunction and Improves Therapeutic Response in PI3K alpha-Mutant Xenografts.
Cancer Discov, 13, 2023
|
|
3LRU
 
 | |
1QJL
 
 | | METALLOTHIONEIN MTA FROM SEA URCHIN (BETA DOMAIN) | | Descriptor: | CADMIUM ION, METALLOTHIONEIN | | Authors: | Riek, R, Precheur, B, Wang, Y, Mackay, E.A, Wider, G, Guntert, P, Liu, A, Kaegi, J.H.R, Wuthrich, K. | | Deposit date: | 1999-06-24 | | Release date: | 1999-08-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the sea urchin (Strongylocentrotus purpuratus) metallothionein MTA.
J. Mol. Biol., 291, 1999
|
|
4K1S
 
 | | Gly-Ser-SplB protease from Staphylococcus aureus at 1.96 A resolution | | Descriptor: | Serine protease SplB | | Authors: | Zdzalik, M, Pustelny, K, Stec-Niemczyk, J, Cichon, P, Czarna, A, Popowicz, G, Drag, M, Wladyka, B, Potempa, J, Dubin, A, Dubin, G. | | Deposit date: | 2013-04-05 | | Release date: | 2014-04-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Staphylococcal SplB Serine Protease Utilizes a Novel Molecular Mechanism of Activation.
J.Biol.Chem., 289, 2014
|
|
6DFL
 
 | | WaaP in complex with acyl carrier protein | | Descriptor: | Acyl carrier protein, Lipopolysaccharide core heptose(I) kinase RfaP, S-[2-({N-[(2S)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alanyl}amino)ethyl] hexadecanethioate | | Authors: | Chopra, R, Vash, B. | | Deposit date: | 2018-05-15 | | Release date: | 2019-04-03 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (2.396 Å) | | Cite: | Acylated-acyl carrier protein stabilizes the Pseudomonas aeruginosa WaaP lipopolysaccharide heptose kinase.
Sci Rep, 8, 2018
|
|
3LJF
 
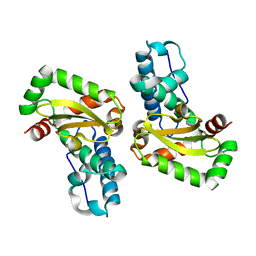 | | The X-ray structure of iron superoxide dismutase from Pseudoalteromonas haloplanktis (crystal form II) | | Descriptor: | FE (III) ION, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose, iron superoxide dismutase | | Authors: | Merlino, A, Russo Krauss, I, Rossi, B, Conte, M, Vergara, A, Sica, F. | | Deposit date: | 2010-01-26 | | Release date: | 2010-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and flexibility in cold-adapted iron superoxide dismutases: the case of the enzyme isolated from Pseudoalteromonas haloplanktis.
J.Struct.Biol., 172, 2010
|
|
2AI9
 
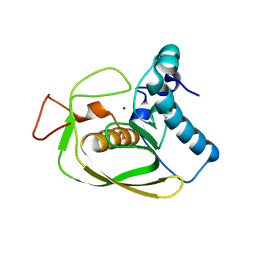 | | S.aureus Polypeptide Deformylase | | Descriptor: | NICKEL (II) ION, Peptide deformylase, SULFATE ION | | Authors: | Smith, K.J, Petit, C.M, Aubart, K, Smyth, M, McManus, E, Jones, J, Fosberry, A, Lewis, C, Lonetto, M, Christensen, S.B. | | Deposit date: | 2005-07-29 | | Release date: | 2005-09-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Variation and inhibitor binding in polypeptide deformylase from four different bacterial species.
Protein Sci., 12, 2003
|
|
1QMN
 
 | |
4JZD
 
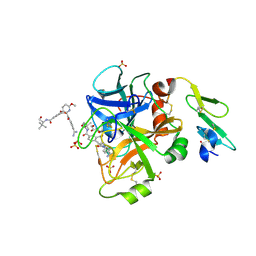 | | Structure of factor VIIA in complex with the inhibitor 2-{2-[(4-carbamimidoylphenyl)carbamoyl]-6-methoxypyridin-3-yl}-5-{[(2S)-1-hydroxy-3,3-dimethylbutan-2-yl]carbamoyl}benzoic acid | | Descriptor: | 2-{2-[(4-carbamimidoylphenyl)carbamoyl]-6-methoxypyridin-3-yl}-5-{[(2S)-1-hydroxy-3,3-dimethylbutan-2-yl]carbamoyl}benzoic acid, CALCIUM ION, Factor VIIa (Heavy Chain), ... | | Authors: | Jacobson, B.L, Anumula, R. | | Deposit date: | 2013-04-02 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of nonbenzamidine factor VIIa inhibitors using a biaryl acid scaffold.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
8SE5
 
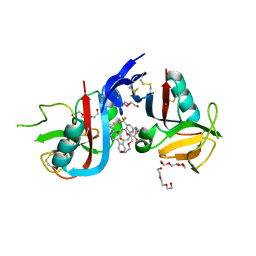 | | NKG2D complexed with inhibitor 14 | | Descriptor: | DI(HYDROXYETHYL)ETHER, N-{(1S)-2-[(1R,5S,6S)-6-(hydroxymethyl)-3-azabicyclo[3.1.0]hexan-3-yl]-2-oxo-1-[3-(trifluoromethyl)phenyl]ethyl}-4'-(trifluoromethyl)[1,1'-biphenyl]-2-carboxamide, NKG2-D type II integral membrane protein, ... | | Authors: | Thompson, A.A, Grant, J.C, Karpowich, N.K, Sharma, S. | | Deposit date: | 2023-04-08 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Development of small molecule inhibitors of natural killer group 2D receptor (NKG2D).
Bioorg.Med.Chem.Lett., 96, 2023
|
|
4BD3
 
 | |
3LYX
 
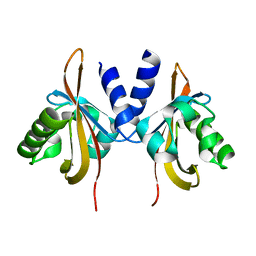 | | Crystal structure of the PAS domain of the protein CPS_1291 from Colwellia psychrerythraea. Northeast Structural Genomics Consortium target id CsR222B | | Descriptor: | Sensory box/GGDEF domain protein | | Authors: | Seetharaman, J, Chen, Y, Wang, D, Janjua, H, Cunningham, K, Owens, L, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-02-28 | | Release date: | 2010-04-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the PAS domain of the protein CPS_1291 from Colwellia psychrerythraea. Northeast Structural Genomics Consortium target id CsR222B
To be Published
|
|
6DGF
 
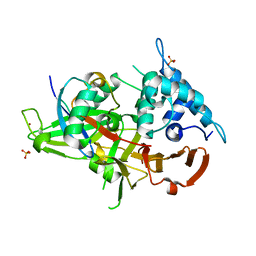 | | Ubiquitin Variant bound to USP2 | | Descriptor: | Polyubiquitin-B, SULFATE ION, Ubiquitin carboxyl-terminal hydrolase 2, ... | | Authors: | Manczyk, N, Sicheri, F. | | Deposit date: | 2018-05-17 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Yeast Two-Hybrid Analysis for Ubiquitin Variant Inhibitors of Human Deubiquitinases.
J. Mol. Biol., 431, 2019
|
|
6WTM
 
 | |
3LX8
 
 | | Crystal structure of GDP-bound NFeoB from S. thermophilus | | Descriptor: | Ferrous iron uptake transporter protein B, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Ash, M.R, Guilfoyle, A, Maher, M.J, Clarke, R.J, Guss, J.M, Jormakka, M. | | Deposit date: | 2010-02-24 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Potassium-activated GTPase reaction in the G Protein-coupled ferrous iron transporter B.
J.Biol.Chem., 285, 2010
|
|
6DM8
 
 | | Understanding the Species Selectivity of Myeloid cell leukemia-1 (Mcl-1) inhibitors | | Descriptor: | 4-{8-chloro-11-[3-(4-chloro-3,5-dimethylphenoxy)propyl]-1-oxo-7-(1,3,5-trimethyl-1H-pyrazol-4-yl)-4,5-dihydro-1H-[1,4]diazepino[1,2-a]indol-2(3H)-yl}-1-methyl-1H-indole-6-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 homolog - MBP chimera, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Zhao, B. | | Deposit date: | 2018-06-04 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Understanding the Species Selectivity of Myeloid Cell Leukemia-1 (Mcl-1) Inhibitors.
Biochemistry, 57, 2018
|
|
1QAP
 
 | | QUINOLINIC ACID PHOSPHORIBOSYLTRANSFERASE WITH BOUND QUINOLINIC ACID | | Descriptor: | QUINOLINIC ACID, QUINOLINIC ACID PHOSPHORIBOSYLTRANSFERASE | | Authors: | Eads, J.C, Ozturk, D, Wexler, T.B, Grubmeyer, C, Sacchettini, J.C. | | Deposit date: | 1996-09-20 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A new function for a common fold: the crystal structure of quinolinic acid phosphoribosyltransferase.
Structure, 5, 1997
|
|
