4LPB
 
 | | Crystal structure of a topoisomerase ATPase inhibitor | | Descriptor: | 1-ethyl-3-{5'-(5-oxo-4,5-dihydro-1,3,4-oxadiazol-2-yl)-4-[4-(trifluoromethyl)-1,3-thiazol-2-yl]-3,3'-bipyridin-6-yl}urea, Topoisomerase IV subunit B | | Authors: | Boriack-Sjodin, A. | | Deposit date: | 2013-07-15 | | Release date: | 2013-10-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Fragment-to-Hit-to-Lead Discovery of a Novel Pyridylurea Scaffold of ATP Competitive Dual Targeting Type II Topoisomerase Inhibiting Antibacterial Agents.
J.Med.Chem., 56, 2013
|
|
2XD6
 
 | | Hsp90 complexed with a resorcylic acid macrolactone. | | Descriptor: | (5Z)-13-CHLORO-14,16-DIHYDROXY-1,11-DIOXO-3,4,7,8,9,10,11,12-OCTAHYDRO-1H-2-BENZOXACYCLOTETRADECINE-6-CARBALDEHYDE, ATP-DEPENDENT MOLECULAR CHAPERONE HSP82, GLYCEROL | | Authors: | Roe, S.M, Prodromou, C, Pearl, L.H, Moody, C.J. | | Deposit date: | 2010-04-29 | | Release date: | 2010-08-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Inhibition of Hsp90 with Resorcylic Acid Macrolactones. Synthesis and Binding Studies.
Chemistry, 16, 2010
|
|
3EHG
 
 | |
4DUH
 
 | | Crystal structure of 24 kDa domain of E. coli DNA gyrase B in complex with small molecule inhibitor | | Descriptor: | 4-{[4'-methyl-2'-(propanoylamino)-4,5'-bi-1,3-thiazol-2-yl]amino}benzoic acid, DNA gyrase subunit B | | Authors: | Brvar, M, Renko, M, Perdih, A, Solmajer, T, Turk, D. | | Deposit date: | 2012-02-22 | | Release date: | 2012-08-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-based discovery of substituted 4,5'-bithiazoles as novel DNA gyrase inhibitors.
J.Med.Chem., 55, 2012
|
|
8H70
 
 | |
5MMP
 
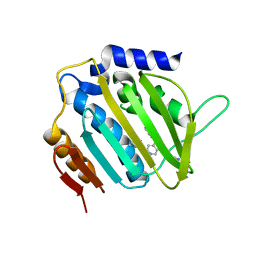 | | E. coli DNA Gyrase B 24 kDa ATPase domain in complex with 1-ethyl-3-[5-pyridin-4-yl-8-(pyridin-3-ylamino)-isoquinolin-3-yl]-urea | | Descriptor: | 1-ethyl-3-[5-pyridin-4-yl-8-(pyridin-3-ylamino)isoquinolin-3-yl]urea, DNA gyrase subunit B | | Authors: | Panchaud, P, Bruyere, T, Blumstein, A.-C, Bur, D, Chambovey, A, Ertel, E.A, Gude, M, Hubschwerlen, C, Jacob, L, Kimmerlin, T, Pfeifer, T, Prade, L, Seiler, P, Ritz, D, Rueedi, G. | | Deposit date: | 2016-12-12 | | Release date: | 2017-04-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery and Optimization of Isoquinoline Ethyl Ureas as Antibacterial Agents.
J. Med. Chem., 60, 2017
|
|
4P8O
 
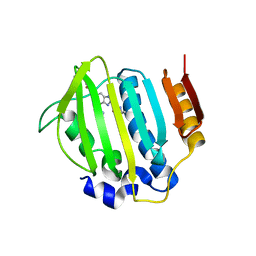 | | S. aureus gyrase bound to an aminobenzimidazole urea inhibitor | | Descriptor: | 1-ethyl-3-[5-(5-fluoropyridin-3-yl)-7-(pyrimidin-2-yl)-1H-benzimidazol-2-yl]urea, DNA gyrase subunit B | | Authors: | Jacobs, M.D. | | Deposit date: | 2014-03-31 | | Release date: | 2014-10-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Second-generation antibacterial benzimidazole ureas: discovery of a preclinical candidate with reduced metabolic liability.
J.Med.Chem., 57, 2014
|
|
4PL9
 
 | |
5MMO
 
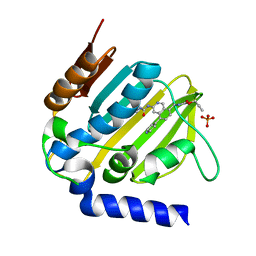 | | E. coli DNA Gyrase B 24 kDa ATPase domain in complex with [3-(3-ethyl-ureido)-5-(pyridin-4-yl)-isoquinolin-8-yl-methyl]-carbamic acid prop-2-ynyl ester | | Descriptor: | DNA gyrase subunit B, PHOSPHATE ION, prop-2-ynyl ~{N}-[[3-(ethylcarbamoylamino)-5-pyridin-4-yl-isoquinolin-8-yl]methyl]carbamate | | Authors: | Panchaud, P, Bruyere, T, Blumstein, A.-C, Bur, D, Chambovey, A, Ertel, E.A, Gude, M, Hubschwerlen, C, Jacob, L, Kimmerlin, T, Pfeifer, T, Prade, L, Seiler, P, Ritz, D, Rueedi, G. | | Deposit date: | 2016-12-12 | | Release date: | 2017-04-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Discovery and Optimization of Isoquinoline Ethyl Ureas as Antibacterial Agents.
J. Med. Chem., 60, 2017
|
|
5MMN
 
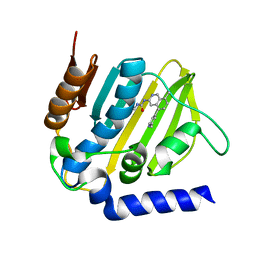 | | E. coli DNA Gyrase B 24 kDa ATPase domain in complex with 1-ethyl-3-[8-methyl-5-(2-methyl-pyridin-4-yl)-isoquinolin-3-yl]-urea | | Descriptor: | 1-ethyl-3-[8-methyl-5-(2-methylpyridin-4-yl)isoquinolin-3-yl]urea, DNA gyrase subunit B | | Authors: | Panchaud, P, Bruyere, T, Blumstein, A.-C, Bur, D, Chambovey, A, Ertel, E.A, Gude, M, Hubschwerlen, C, Jacob, L, Kimmerlin, T, Pfeifer, T, Prade, L, Seiler, P, Ritz, D, Rueedi, G. | | Deposit date: | 2016-12-12 | | Release date: | 2017-04-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and Optimization of Isoquinoline Ethyl Ureas as Antibacterial Agents.
J. Med. Chem., 60, 2017
|
|
3CGZ
 
 | |
4GFN
 
 | | Pyrrolopyrimidine inhibitors of dna gyrase b and topoisomerase iv, part i: structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic | | Descriptor: | (1R,5S,6s)-3-[5-chloro-6-ethyl-2-(pyrimidin-5-ylsulfanyl)-7H-pyrrolo[2,3-d]pyrimidin-4-yl]-3-azabicyclo[3.1.0]hexan-6-amine, DNA gyrase subunit B, GLYCEROL | | Authors: | Bensen, D.C, Trzoss, M, Tari, L.W. | | Deposit date: | 2012-08-03 | | Release date: | 2013-02-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Pyrrolopyrimidine inhibitors of DNA gyrase B (GyrB) and topoisomerase IV (ParE). Part I: Structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4GEE
 
 | | Pyrrolopyrimidine inhibitors of DNA gyrase B and topoisomerase IV, part I: structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity. | | Descriptor: | (1R,5S,6s)-3-[5-chloro-6-ethyl-2-(pyrimidin-5-yloxy)-7H-pyrrolo[2,3-d]pyrimidin-4-yl]-3-azabicyclo[3.1.0]hexan-6-amine, DNA gyrase subunit B, GLYCEROL | | Authors: | Bensen, D.C, Chen, Z, Tari, L.W. | | Deposit date: | 2012-08-01 | | Release date: | 2013-02-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Pyrrolopyrimidine inhibitors of DNA gyrase B (GyrB) and topoisomerase IV (ParE). Part I: Structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3G7B
 
 | |
3FV5
 
 | |
5IDM
 
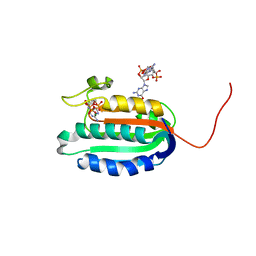 | | Bifunctional histidine kinase CckA (domain, CA) in complex with c-di-GMP and AMPPNP/Mg2+ | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Cell cycle histidine kinase CckA, MAGNESIUM ION, ... | | Authors: | Dubey, B.N, Schirmer, T. | | Deposit date: | 2016-02-24 | | Release date: | 2016-10-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cyclic di-GMP mediates a histidine kinase/phosphatase switch by noncovalent domain cross-linking.
Sci Adv, 2, 2016
|
|
3G75
 
 | |
3G7E
 
 | |
3H80
 
 | | Crystal structure of the amino-terminal domain of HSP90 from Leishmania major, LmjF33.0312:M1-K213 | | Descriptor: | 1,2-ETHANEDIOL, Heat shock protein 83-1, MAGNESIUM ION, ... | | Authors: | Wernimont, A.K, Tempel, W, Lin, Y.H, Hutchinson, A, Mackenzie, F, Fairlamb, A, Kozieradzki, I, Cossar, D, Zhao, Y, Schapira, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Ferguson, M.A.J, Hui, R, Pizarro, J.C, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-04-28 | | Release date: | 2009-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the amino-terminal domain of HSP90 from Leishmania major, LmjF33.0312:M1-K213
To be Published
|
|
9MT9
 
 | |
9MSV
 
 | |
9MSQ
 
 | |
9MT3
 
 | | Candida albicans Hsp90 nucleotide binding domain in complex with XL888 | | Descriptor: | 5-[(2R)-butan-2-ylamino]-N-{(3-endo)-8-[5-(cyclopropylcarbonyl)pyridin-2-yl]-8-azabicyclo[3.2.1]oct-3-yl}-2-methylbenzene-1,4-dicarboxamide, Heat shock protein 90 homolog | | Authors: | Kowalewski, M.E, Redinbo, M.R. | | Deposit date: | 2025-01-10 | | Release date: | 2025-06-04 | | Last modified: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural Insights into Selectively Targeting Candida albicans Hsp90.
Biochemistry, 64, 2025
|
|
9MSX
 
 | | Candida albicans Hsp90 nucleotide binding domain in complex with TAS116 | | Descriptor: | (4P)-3-ethyl-4-[(4P)-4-[(4M)-4-(1-methyl-1H-pyrazol-4-yl)-1H-imidazol-1-yl]-3-(propan-2-yl)-1H-pyrazolo[3,4-b]pyridin-1-yl]benzamide, 1,2-ETHANEDIOL, Heat shock protein 90 homolog | | Authors: | Kowalewski, M.E, Redinbo, M.R. | | Deposit date: | 2025-01-10 | | Release date: | 2025-06-04 | | Last modified: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Insights into Selectively Targeting Candida albicans Hsp90.
Biochemistry, 64, 2025
|
|
9MTB
 
 | |
