5NAV
 
 | |
2O9P
 
 | | beta-glucosidase B from Paenibacillus polymyxa | | Descriptor: | Beta-glucosidase B | | Authors: | Isorna, P, Polaina, J, Sanz-Aparicio, J. | | Deposit date: | 2006-12-14 | | Release date: | 2007-10-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of Paenibacillus polymyxa beta-Glucosidase B Complexes Reveal the Molecular Basis of Substrate Specificity and Give New Insights into the Catalytic Machinery of Family I Glycosidases
J.Mol.Biol., 371, 2007
|
|
7BZM
 
 | | Crystal structure of rice Os3BGlu7 with glucoimidazole | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-glucosidase 7, GLUCOIMIDAZOLE, ... | | Authors: | Pengthaisong, S, Ketudat Cairns, J.R, Tankrathok, A. | | Deposit date: | 2020-04-28 | | Release date: | 2020-07-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of Specific Glucoimidazole and Mannoimidazole Binding by Os3BGlu7.
Biomolecules, 10, 2020
|
|
4EAN
 
 | | 1.75A resolution structure of indole bound beta-glycosidase (W33G) from sulfolobus solfataricus | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Beta-galactosidase, CHLORIDE ION, ... | | Authors: | Lovell, S, Battaile, K.P, Deckert, K, Brunner, L.C, Budiardjo, S.J, Karanicolas, J. | | Deposit date: | 2012-03-22 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Designing allosteric control into enzymes by chemical rescue of structure.
J.Am.Chem.Soc., 134, 2012
|
|
4EK7
 
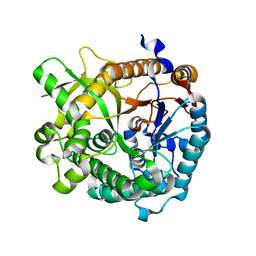 | | High speed X-ray analysis of plant enzymes at room temperature | | Descriptor: | CHLORIDE ION, Raucaffricine-O-beta-D-glucosidase, beta-D-glucopyranose | | Authors: | Xia, L, Rajendran, C, Ruppert, M, Panjikar, S, Wang, M, Stoeckigt, J. | | Deposit date: | 2012-04-09 | | Release date: | 2013-02-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | High speed X-ray analysis of plant enzymes at room temperature.
Phytochemistry, 2012
|
|
4EAM
 
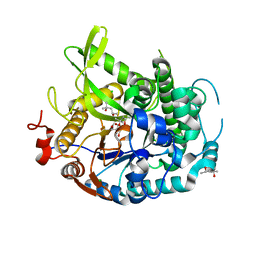 | | 1.70A resolution structure of apo beta-glycosidase (W33G) from sulfolobus solfataricus | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-galactosidase, ... | | Authors: | Lovell, S, Battaile, K.P, Deckert, K, Brunner, L.C, Budiardjo, S.J, Karanicolas, J. | | Deposit date: | 2012-03-22 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Designing allosteric control into enzymes by chemical rescue of structure.
J.Am.Chem.Soc., 134, 2012
|
|
7D6B
 
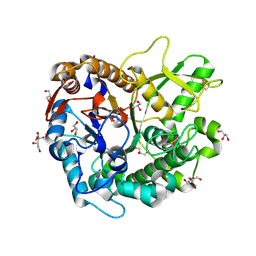 | | Crystal structure of Oryza sativa Os4BGlu18 monolignol beta-glucosidase with delta-gluconolactone | | Descriptor: | Beta-glucosidase 18, D-glucono-1,5-lactone, GLYCEROL, ... | | Authors: | Baiya, S, Pengthaisong, S, Ketudat Cairns, J.R. | | Deposit date: | 2020-09-29 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural analysis of rice Os4BGlu18 monolignol beta-glucosidase.
Plos One, 16, 2021
|
|
7D6A
 
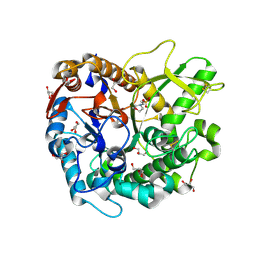 | | Crystal structure of Oryza sativa Os4BGlu18 monolignol beta-glucosidase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-glucosidase 18, GLYCEROL, ... | | Authors: | Baiya, S, Pengthaisong, S, Ketudat Cairns, J.R. | | Deposit date: | 2020-09-29 | | Release date: | 2021-01-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural analysis of rice Os4BGlu18 monolignol beta-glucosidase.
Plos One, 16, 2021
|
|
4ZE4
 
 | | Structure of Gan1D, a putative 6-phospho-beta-galactosidase from Geobacillus stearothermophilus | | Descriptor: | GLYCEROL, IMIDAZOLE, Putative 6-phospho-beta-galactobiosidase | | Authors: | Lansky, S, Zehavi, A, Dvir, H, Shoham, Y, Shoham, G. | | Deposit date: | 2015-04-20 | | Release date: | 2016-06-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structure of Gan1D, a putative 6-phospho-beta-galactosidase from Geobacillus stearothermophilus
To Be Published
|
|
4ZFM
 
 | | Structure of Gan1D-E170Q in complex with cellobiose-6-phosphate | | Descriptor: | 1,5-anhydro-6-O-phosphono-D-glucitol, 1,5-anhydro-6-O-phosphono-D-glucitol-(1-4)-beta-D-glucopyranose, 6-O-phosphono-beta-D-glucopyranose, ... | | Authors: | Lansky, S, Zehavi, A, Dvir, H, Shoham, Y, Shoham, G. | | Deposit date: | 2015-04-21 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of Gan1D-E170Q in complex with cellobiose-6-phosphate
To Be Published
|
|
4ZEN
 
 | | Structure of Gan1D, a putative 6-phospho-beta-galactosidase from Geobacillus stearothermophilus, in complex with 6-phospho-beta-galactose | | Descriptor: | 6-O-phosphono-beta-D-galactopyranose, GLYCEROL, IMIDAZOLE, ... | | Authors: | Lansky, S, Zehavi, A, Dvir, H, Shoham, Y, Shoham, G. | | Deposit date: | 2015-04-20 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure of Gan1D, a putative 6-phospho-beta-galactosidase from Geobacillus stearothermophilus, in complex with 6-phospho-beta-galactose
To Be Published
|
|
4ZEP
 
 | | Structure of Gan1D, a 6-phospho-beta-galactosidase from Geobacillus stearothermophilus, in complex with 6-phospho-glucose | | Descriptor: | 6-O-phosphono-beta-D-glucopyranose, GLYCEROL, IMIDAZOLE, ... | | Authors: | Lansky, S, Zehavi, A, Dvir, H, Shoham, Y, Shoham, G. | | Deposit date: | 2015-04-20 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structure of Gan1D, a 6-phospho-beta-galactosidase from Geobacillus stearothermophilus, in complex with 6-phospho-glucose
To Be Published
|
|
3PBG
 
 | | 6-PHOSPHO-BETA-GALACTOSIDASE FORM-C | | Descriptor: | 6-PHOSPHO-BETA-D-GALACTOSIDASE, SULFATE ION | | Authors: | Wiesmann, C, Schulz, G.E. | | Deposit date: | 1997-02-21 | | Release date: | 1997-07-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures and mechanism of 6-phospho-beta-galactosidase from Lactococcus lactis.
J.Mol.Biol., 269, 1997
|
|
4ZE5
 
 | | Structure of Gan1D-E170Q, a catalytic mutant of a putative 6-phospho-beta-galactosidase from Geobacillus stearothermophilus | | Descriptor: | GLYCEROL, IMIDAZOLE, Putative 6-phospho-beta-galactobiosidase | | Authors: | Lansky, S, Zehavi, A, Dvir, H, Shoham, Y, Shoham, G. | | Deposit date: | 2015-04-20 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structure of Gan1D-E170Q, a catalytic mutant of a putative 6-phospho-beta-galactosidase from Geobacillus stearothermophilus
To Be Published
|
|
4F66
 
 | | The crystal structure of 6-phospho-beta-glucosidase from Streptococcus mutans UA159 in complex with beta-D-glucose-6-phosphate. | | Descriptor: | 1,2-ETHANEDIOL, 6-O-phosphono-beta-D-glucopyranose, FORMIC ACID, ... | | Authors: | Tan, K, Michalska, K, Hatzos-Skintges, C, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-05-14 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.479 Å) | | Cite: | GH1-family 6-P-beta-glucosidases from human microbiome lactic acid bacteria.
Acta Crystallogr. D Biol. Crystallogr., 69, 2013
|
|
3PTK
 
 | | The crystal structure of rice (Oryza sativa L.) Os4BGlu12 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-glucosidase Os4BGlu12, ZINC ION | | Authors: | Sansenya, S, Opassiri, R, Kuaprasert, B, Chen, C.J, Ketudat Cairns, J.R. | | Deposit date: | 2010-12-03 | | Release date: | 2011-05-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | The crystal structure of rice (Oryza sativa L.) Os4BGlu12, an oligosaccharide and tuberonic acid glucoside-hydrolyzing beta-glucosidase with significant thioglucohydrolase activity
Arch.Biochem.Biophys., 510, 2011
|
|
3PTM
 
 | | The crystal structure of rice (Oryza sativa L.) Os4BGlu12 with 2-fluoroglucopyranoside | | Descriptor: | 2-deoxy-2-fluoro-alpha-D-glucopyranose, Beta-glucosidase Os4BGlu12, GLYCEROL, ... | | Authors: | Sansenya, S, Opassiri, R, Kuaprasert, B, Chen, C.J, Ketudat Cairns, J.R. | | Deposit date: | 2010-12-03 | | Release date: | 2011-05-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of rice (Oryza sativa L.) Os4BGlu12, an oligosaccharide and tuberonic acid glucoside-hydrolyzing beta-glucosidase with significant thioglucohydrolase activity
Arch.Biochem.Biophys., 510, 2011
|
|
4F79
 
 | | The crystal structure of 6-phospho-beta-glucosidase mutant (E375Q) in complex with Salicin 6-phosphate | | Descriptor: | 2-(hydroxymethyl)phenyl 6-O-phosphono-beta-D-glucopyranoside, GLYCEROL, Putative phospho-beta-glucosidase | | Authors: | Tan, K, Michalska, K, Li, H, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-05-15 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | GH1-family 6-P-beta-glucosidases from human microbiome lactic acid bacteria.
Acta Crystallogr. D Biol. Crystallogr., 69, 2013
|
|
7BBS
 
 | |
3PTQ
 
 | | The crystal structure of rice (Oryza sativa L.) Os4BGlu12 with dinitrophenyl 2-deoxy-2-fluoro-beta-D-glucopyranoside | | Descriptor: | 2,4-dinitrophenyl 2-deoxy-2-fluoro-beta-D-glucopyranoside, Beta-glucosidase Os4BGlu12, GLYCEROL, ... | | Authors: | Sansenya, S, Opassiri, R, Kuaprasert, B, Chen, C.J, Ketudat Cairns, J.R. | | Deposit date: | 2010-12-03 | | Release date: | 2011-05-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The crystal structure of rice (Oryza sativa L.) Os4BGlu12, an oligosaccharide and tuberonic acid glucoside-hydrolyzing beta-glucosidase with significant thioglucohydrolase activity
Arch.Biochem.Biophys., 510, 2011
|
|
7E5J
 
 | | Crystal structure of beta-glucosidase from Thermoanaerobacterium saccharolyticum | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-glucosidase, SODIUM ION | | Authors: | Nam, K.H. | | Deposit date: | 2021-02-18 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Biochemical and Structural Analysis of a Glucose-Tolerant beta-Glucosidase from the Hemicellulose-Degrading Thermoanaerobacterium saccharolyticum.
Molecules, 27, 2022
|
|
3PN8
 
 | | The crystal structure of 6-phospho-beta-glucosidase from Streptococcus mutans UA159 | | Descriptor: | DI(HYDROXYETHYL)ETHER, FORMIC ACID, Putative phospho-beta-glucosidase, ... | | Authors: | Tan, K, Li, H, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-11-18 | | Release date: | 2010-12-15 | | Last modified: | 2012-08-08 | | Method: | X-RAY DIFFRACTION (1.693 Å) | | Cite: | The crystal structure of 6-phospho-beta-glucosidase from Streptococcus mutants UA159
To be Published
|
|
6KHT
 
 | | Chimeric beta-glucosidase Cel1b-H13 | | Descriptor: | Glycoside hydrolase family 1 | | Authors: | Niu, K.L. | | Deposit date: | 2019-07-16 | | Release date: | 2021-01-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.305 Å) | | Cite: | Chimeric beta-glucosidase Cel1b-H13
To Be Published
|
|
2WBG
 
 | | Structure of family 1 beta-glucosidase from Thermotoga maritima in complex with 3-imino-2-oxa-(+)-castanospermine | | Descriptor: | (3Z,5S,6R,7S,8R,8aR)-3-(octylimino)hexahydro[1,3]oxazolo[3,4-a]pyridine-5,6,7,8-tetrol, ACETATE ION, BETA-GLUCOSIDASE A | | Authors: | Aguilar, M, Gloster, T.M, Turkenburg, J.P, Garcia-Moreno, M.I, Ortiz Mellet, C, Davies, G.J, Garcia Fernandez, J.M. | | Deposit date: | 2009-02-27 | | Release date: | 2009-04-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Glycosidase Inhibition by Ring-Modified Castanospermine Analogues: Tackling Enzyme Selectivity by Inhibitor Tailoring.
Org.Biomol.Chem., 7, 2009
|
|
6QWI
 
 | | Structure of beta-glucosidase A from Paenibacillus polymyxa complexed with multivalent inhibitors. | | Descriptor: | (2~{S},3~{S},4~{R})-2-[[4-[4-(2-ethoxyethoxy)phenyl]-1,2,3-triazol-1-yl]methyl]pyrrolidine-3,4-diol, Beta-glucosidase A | | Authors: | Jimenez-Ortega, E, Sanz-Aparicio, J. | | Deposit date: | 2019-03-05 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural basis of the inhibition of GH1 beta-glucosidases by multivalent pyrrolidine iminosugars.
Bioorg.Chem., 89, 2019
|
|
