1AYU
 
 | | CRYSTAL STRUCTURE OF CYSTEINE PROTEASE HUMAN CATHEPSIN K IN COMPLEX WITH A COVALENT SYMMETRIC BISCARBOHYDRAZIDE INHIBITOR | | Descriptor: | 1,5-BIS(N-BENZYLOXYCARBONYL-L-LEUCINYL)CARBOHYDRAZIDE, CATHEPSIN K | | Authors: | Zhao, B, Smith, W.W, Janson, C.A, Abdel-Meguid, S.S. | | Deposit date: | 1997-11-10 | | Release date: | 1998-11-25 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design of potent and selective human cathepsin K inhibitors that span the active site.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
1AYW
 
 | | CRYSTAL STRUCTURE OF CYSTEINE PROTEASE HUMAN CATHEPSIN K IN COMPLEX WITH A COVALENT BENZYLOXYBENZOYLCARBOHYDRAZIDE INHIBITOR | | Descriptor: | 1-(N-BENZYLOXYCARBONYL-L-LEUCINYL)-5-(3-BENZYLOXY BENZOYL)CARBOHYDRAZIDE, CATHEPSIN K | | Authors: | Zhao, B, Smith, W.W, Janson, C.A, Abdel-Meguid, S.S. | | Deposit date: | 1997-11-10 | | Release date: | 1998-11-25 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design of potent and selective human cathepsin K inhibitors that span the active site.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
254L
 
 | | LYSOZYME | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, LYSOZYME | | Authors: | Kuroki, R, Shoichet, B, Weaver, L.H, Matthews, B.W. | | Deposit date: | 1997-11-10 | | Release date: | 1998-01-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A relationship between protein stability and protein function.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
255L
 
 | | HYDROLASE | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, LYSOZYME | | Authors: | Kuroki, R, Shoichet, B, Weaver, L.H, Matthews, B.W. | | Deposit date: | 1997-11-10 | | Release date: | 1998-01-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A relationship between protein stability and protein function.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
253L
 
 | | LYSOZYME | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, LYSOZYME | | Authors: | Kuroki, R, Shoichet, B, Weaver, L.H, Matthews, B.W. | | Deposit date: | 1997-11-10 | | Release date: | 1998-01-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A relationship between protein stability and protein function.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
1KIM
 
 | | CRYSTAL STRUCTURE OF THYMIDINE KINASE FROM HERPES SIMPLEX VIRUS TYPE I COMPLEXED WITH DEOXYTHYMIDINE | | Descriptor: | SULFATE ION, THYMIDINE, THYMIDINE KINASE | | Authors: | Champness, J.N, Bennett, M.S, Wien, F, Brown, D.G, Visse, R, Sandhu, G, Davies, A, Rizkallah, P.J, Melitz, C, Summers, W.C, Sanderson, M.R. | | Deposit date: | 1997-11-12 | | Release date: | 1998-05-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Exploring the active site of herpes simplex virus type-1 thymidine kinase by X-ray crystallography of complexes with aciclovir and other ligands.
Proteins, 32, 1998
|
|
1AYX
 
 | | CRYSTAL STRUCTURE OF GLUCOAMYLASE FROM SACCHAROMYCOPSIS FIBULIGERA AT 1.7 ANGSTROMS | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLUCOAMYLASE | | Authors: | Sevcik, J, Hostinova, E, Gasperik, J, Solovicova, A, Wilson, K.S, Dauter, Z. | | Deposit date: | 1997-11-12 | | Release date: | 1998-05-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of glucoamylase from Saccharomycopsis fibuligera at 1.7 A resolution.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1AYZ
 
 | |
1AYY
 
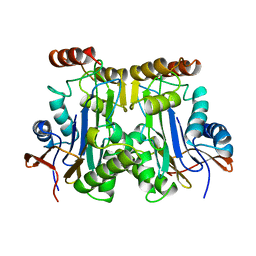 | | GLYCOSYLASPARAGINASE | | Descriptor: | GLYCOSYLASPARAGINASE | | Authors: | Van Roey, P, Xuan, J. | | Deposit date: | 1997-11-12 | | Release date: | 1998-04-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Crystal structure of glycosylasparaginase from Flavobacterium meningosepticum.
Protein Sci., 7, 1998
|
|
2H1P
 
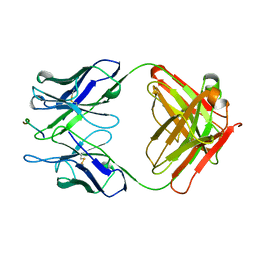 | | THE THREE-DIMENSIONAL STRUCTURES OF A POLYSACCHARIDE BINDING ANTIBODY TO CRYPTOCOCCUS NEOFORMANS AND ITS COMPLEX WITH A PEPTIDE FROM A PHAGE DISPLAY LIBRARY: IMPLICATIONS FOR THE IDENTIFICATION OF PEPTIDE MIMOTOPES | | Descriptor: | 2H1, PA1 | | Authors: | Young, A.C.M, Valadon, P, Casadevall, A, Scharff, M.D, Sacchettini, J.C. | | Deposit date: | 1997-11-12 | | Release date: | 1998-01-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The three-dimensional structures of a polysaccharide binding antibody to Cryptococcus neoformans and its complex with a peptide from a phage display library: implications for the identification of peptide mimotopes.
J.Mol.Biol., 274, 1997
|
|
1AY0
 
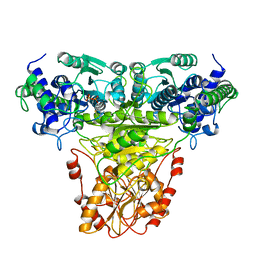 | | IDENTIFICATION OF CATALYTICALLY IMPORTANT RESIDUES IN YEAST TRANSKETOLASE | | Descriptor: | CALCIUM ION, THIAMINE DIPHOSPHATE, TRANSKETOLASE | | Authors: | Wikner, C, Nilsson, U, Meshalkina, L, Udekwu, C, Lindqvist, Y, Schneider, G. | | Deposit date: | 1997-11-13 | | Release date: | 1998-05-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification of catalytically important residues in yeast transketolase.
Biochemistry, 36, 1997
|
|
1AY2
 
 | | STRUCTURE OF THE FIBER-FORMING PROTEIN PILIN AT 2.6 ANGSTROMS RESOLUTION | | Descriptor: | HEPTANE-1,2,3-TRIOL, PLATINUM (II) ION, TYPE 4 PILIN, ... | | Authors: | Forest, K.T, Parge, H.E, Tainer, J.A. | | Deposit date: | 1997-11-13 | | Release date: | 1998-04-29 | | Last modified: | 2023-02-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the fibre-forming protein pilin at 2.6 A resolution.
Nature, 378, 1995
|
|
1914
 
 | | SIGNAL RECOGNITION PARTICLE ALU RNA BINDING HETERODIMER, SRP9/14 | | Descriptor: | BETA-MERCAPTOETHANOL, PHOSPHATE ION, SIGNAL RECOGNITION PARTICLE 9/14 FUSION PROTEIN | | Authors: | Birse, D, Kapp, U, Strub, K, Cusack, S, Aberg, A. | | Deposit date: | 1997-11-13 | | Release date: | 1998-12-30 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | The crystal structure of the signal recognition particle Alu RNA binding heterodimer, SRP9/14.
EMBO J., 16, 1997
|
|
1AY1
 
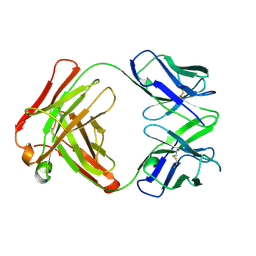 | | ANTI TAQ FAB TP7 | | Descriptor: | TP7 FAB | | Authors: | Murali, R, Helmer-Citterich, M, Sharkey, D.J, Scalice, E.R, Daiss, J.L, Sullivan, M.A, Murthy, H.M.K. | | Deposit date: | 1997-11-13 | | Release date: | 1998-05-13 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural studies on an inhibitory antibody against Thermus aquaticus DNA polymerase suggest mode of inhibition.
Protein Eng., 11, 1998
|
|
1AY6
 
 | | THROMBIN INHIBITOR FROM THEONALLA, CYCLOTHEANAMIDE-BASED MACROCYCLIC TRIPEPTIDE MOTIF | | Descriptor: | HIRUGEN, THROMBIN HEAVY CHAIN, THROMBIN LIGHT CHAIN, ... | | Authors: | Ganesh, V, Maryanoff, B.E, Tulinsky, A. | | Deposit date: | 1997-11-14 | | Release date: | 1998-03-18 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Novel thrombin inhibitors that are based on a macrocyclic tripeptide motif
Bioorg.Med.Chem.Lett., 6, 1996
|
|
1AY4
 
 | | AROMATIC AMINO ACID AMINOTRANSFERASE WITHOUT SUBSTRATE | | Descriptor: | AROMATIC AMINO ACID AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Okamoto, A, Hirotsu, K, Kagamiyama, H. | | Deposit date: | 1997-11-14 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structures of Paracoccus denitrificans aromatic amino acid aminotransferase: a substrate recognition site constructed by rearrangement of hydrogen bond network.
J.Mol.Biol., 280, 1998
|
|
1AY5
 
 | | AROMATIC AMINO ACID AMINOTRANSFERASE COMPLEX WITH MALEATE | | Descriptor: | AROMATIC AMINO ACID AMINOTRANSFERASE, MALEIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Okamoto, A, Hirotsu, K, Kagamiyama, H. | | Deposit date: | 1997-11-14 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of Paracoccus denitrificans aromatic amino acid aminotransferase: a substrate recognition site constructed by rearrangement of hydrogen bond network.
J.Mol.Biol., 280, 1998
|
|
1AY7
 
 | | RIBONUCLEASE SA COMPLEX WITH BARSTAR | | Descriptor: | BARSTAR, GUANYL-SPECIFIC RIBONUCLEASE SA | | Authors: | Sevcik, J, Urbanikova, L, Dauter, Z, Wilson, K.S. | | Deposit date: | 1997-11-14 | | Release date: | 1999-03-02 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Recognition of RNase Sa by the inhibitor barstar: structure of the complex at 1.7 A resolution.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1AY8
 
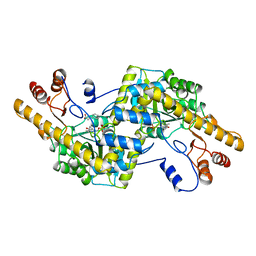 | |
1AY3
 
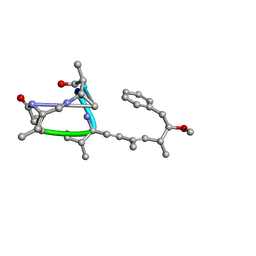 | | Nodularin from Nodularia spumigena | | Descriptor: | PEPTIDIC TOXIN NODULARIN | | Authors: | Annila, A.J. | | Deposit date: | 1997-11-14 | | Release date: | 1999-08-16 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of nodularin. An inhibitor of serine/threonine-specific protein phosphatases.
J.Biol.Chem., 271, 1996
|
|
1SDF
 
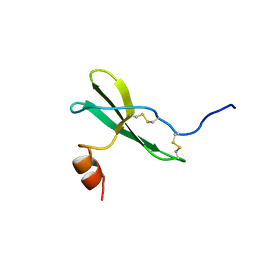 | | SOLUTION STRUCTURE OF STROMAL CELL-DERIVED FACTOR-1 (SDF-1), NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | STROMAL CELL-DERIVED FACTOR-1 | | Authors: | Crump, M.P, Rajarathnam, K, Clark-Lewis, I, Sykes, B.D. | | Deposit date: | 1997-11-15 | | Release date: | 1998-01-28 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure and basis for functional activity of stromal cell-derived factor-1; dissociation of CXCR4 activation from binding and inhibition of HIV-1.
EMBO J., 16, 1997
|
|
1AY9
 
 | | WILD-TYPE UMUD' FROM E. COLI | | Descriptor: | UMUD PROTEIN | | Authors: | Peat, T.S, Frank, E.G, Mcdonald, J.P, Levine, A.S, Woodgate, R, Hendrickson, W.A. | | Deposit date: | 1997-11-15 | | Release date: | 1998-01-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The UmuD' protein filament and its potential role in damage induced mutagenesis.
Structure, 4, 1996
|
|
1GUX
 
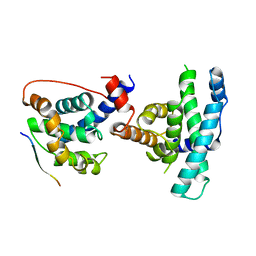 | | RB POCKET BOUND TO E7 LXCXE MOTIF | | Descriptor: | ONCOPROTEIN, RETINOBLASTOMA PROTEIN | | Authors: | Lee, J.O, Russo, A.A, Pavletich, N.P. | | Deposit date: | 1997-11-15 | | Release date: | 1998-12-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of the retinoblastoma tumour-suppressor pocket domain bound to a peptide from HPV E7.
Nature, 391, 1998
|
|
1AZD
 
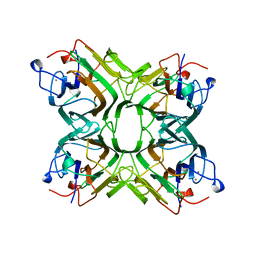 | | CONCANAVALIN FROM CANAVALIA BRASILIENSIS | | Descriptor: | CALCIUM ION, CONBR, MANGANESE (II) ION | | Authors: | Sanz-Aparicio, J, Hermoso, J, Grangeiro, T.B, Calvete, J.J, Cavada, B.S. | | Deposit date: | 1997-11-16 | | Release date: | 1998-04-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of Canavalia brasiliensis lectin suggests a correlation between its quaternary conformation and its distinct biological properties from Concanavalin A.
FEBS Lett., 405, 1997
|
|
4BDP
 
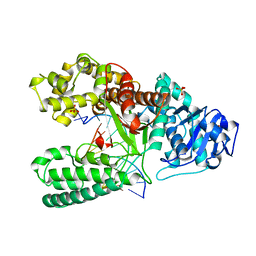 | | CRYSTAL STRUCTURE OF BACILLUS DNA POLYMERASE I FRAGMENT COMPLEXED TO 11 BASE PAIRS OF DUPLEX DNA AFTER ADDITION OF TWO DATP RESIDUES | | Descriptor: | DNA (5'-D(*GP*CP*AP*TP*CP*AP*TP*GP*CP*AP*A)-3'), DNA (5'-D(*TP*AP*TP*TP*GP*CP*AP*TP*GP*AP*TP*GP*C)-3'), MAGNESIUM ION, ... | | Authors: | Kiefer, J.R, Mao, C, Beese, L.S. | | Deposit date: | 1997-11-17 | | Release date: | 1999-01-13 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Visualizing DNA replication in a catalytically active Bacillus DNA polymerase crystal.
Nature, 391, 1998
|
|
