4NTS
 
 | |
7RWB
 
 | | AP2 bound to the APA domain of SGIP in the presence of heparin | | Descriptor: | AP-2 complex subunit alpha-2, AP-2 complex subunit beta, AP-2 complex subunit mu, ... | | Authors: | Baker, R.W, Hollopeter, G, Partlow, E.A. | | Deposit date: | 2021-08-19 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis of an endocytic checkpoint that primes the AP2 clathrin adaptor for cargo internalization.
Nat.Struct.Mol.Biol., 29, 2022
|
|
5FJD
 
 | | APO-CSP1 (COPPER STORAGE PROTEIN 1) FROM METHYLOSINUS TRICHOSPORIUM OB3B | | Descriptor: | COPPER STORAGE PROTEIN 1 | | Authors: | Vita, N, Platsaki, S, Basle, A, Allen, S.J, Paterson, N.G, Crombie, A.T, Murrell, J.C, Waldron, K.J, Dennison, C. | | Deposit date: | 2015-10-07 | | Release date: | 2015-11-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A Four-Helix Bundle Stores Copper for Methane Oxidation.
Nature, 525, 2015
|
|
5FJE
 
 | | CU(I)-CSP1 (COPPER STORAGE PROTEIN 1) FROM METHYLOSINUS TRICHOSPORIUM OB3B | | Descriptor: | COPPER (I) ION, COPPER STORAGE PROTEIN 1, SODIUM ION | | Authors: | Vita, N, Platsaki, S, Basle, A, Allen, S.J, Paterson, N.G, Crombie, A.T, Murrell, J.C, Waldron, K.J, Dennison, C. | | Deposit date: | 2015-10-07 | | Release date: | 2015-11-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Four-Helix Bundle Stores Copper for Methane Oxidation.
Nature, 525, 2015
|
|
4MDH
 
 | |
4MAT
 
 | | E.COLI METHIONINE AMINOPEPTIDASE HIS79ALA MUTANT | | Descriptor: | PROTEIN (METHIONINE AMINOPEPTIDASE), SODIUM ION | | Authors: | Lowther, W.T, Orville, A.M, Madden, D.T, Lim, S, Rich, D.H, Matthews, B.W. | | Deposit date: | 1999-03-29 | | Release date: | 1999-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Escherichia coli methionine aminopeptidase: implications of crystallographic analyses of the native, mutant, and inhibited enzymes for the mechanism of catalysis.
Biochemistry, 38, 1999
|
|
4MKT
 
 | | Human Leukotriene A4 Hydrolase in complex with Pro-Gly-Pro analogue and 4-(4-benzylphenyl)thiazol-2-amine | | Descriptor: | 1-{4-oxo-4-[(2S)-pyrrolidin-2-yl]butanoyl}-L-proline, 4-(4-benzylphenyl)-1,3-thiazol-2-amine, ACETIC ACID, ... | | Authors: | Stsiapanava, A, Rinaldo-Matthis, A, Haeggstrom, J.Z. | | Deposit date: | 2013-09-05 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.618 Å) | | Cite: | Binding of Pro-Gly-Pro at the active site of leukotriene A4 hydrolase/aminopeptidase and development of an epoxide hydrolase selective inhibitor.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4U6Y
 
 | | Crystal Structure of HLA-A*0201 in complex with FLNDK, a 15 mer self-peptide | | Descriptor: | Beta-2-microglobulin, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Gras, S, Chabrol, E, Rossjohn, J. | | Deposit date: | 2014-07-30 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.467 Å) | | Cite: | Naturally Processed Non-canonical HLA-A*02:01 Presented Peptides.
J.Biol.Chem., 290, 2015
|
|
4U6X
 
 | | Crystal Structure of HLA-A*0201 in complex with ALQDA, a 15 mer self-peptide | | Descriptor: | ALQDA peptide, ALQDAGDSSRKEYFI, Beta-2-microglobulin, ... | | Authors: | Gras, S, Chabrol, E, Rossjohn, J. | | Deposit date: | 2014-07-30 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Naturally Processed Non-canonical HLA-A*02:01 Presented Peptides.
J.Biol.Chem., 290, 2015
|
|
3OAB
 
 | | Mint deletion mutant of heterotetrameric geranyl pyrophosphate synthase in complex with ligands | | Descriptor: | 1,2-ETHANEDIOL, 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, DIMETHYLALLYL S-THIOLODIPHOSPHATE, ... | | Authors: | Hsieh, F.-L, Chang, T.-H, Ko, T.-P, Wang, A.H.-J. | | Deposit date: | 2010-08-05 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Enhanced specificity of mint geranyl pyrophosphate synthase by modifying the R-loop interactions
J.Mol.Biol., 404, 2010
|
|
4VGC
 
 | | GAMMA-CHYMOTRYPSIN D-NAPHTHYL-1-ACETAMIDO BORONIC ACID INHIBITOR COMPLEX | | Descriptor: | D-1-NAPHTHYL-2-ACETAMIDO-ETHANE BORONIC ACID, GAMMA CHYMOTRYPSIN, SULFATE ION | | Authors: | Stoll, V.S, Eger, B.T, Hynes, R.C, Martichonok, V, Jones, J.B, Pai, E.F. | | Deposit date: | 1997-05-01 | | Release date: | 1997-11-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Differences in binding modes of enantiomers of 1-acetamido boronic acid based protease inhibitors: crystal structures of gamma-chymotrypsin and subtilisin Carlsberg complexes.
Biochemistry, 37, 1998
|
|
4W96
 
 | | Crystal structure of cross-linked tetragonal hen egg white lysozyme soaked with 5mM [Ru(CO)3Cl2]2 followed by the reaction in deoxy-myoglobin solution | | Descriptor: | CHLORIDE ION, DIMETHYLFORMAMIDE, Lysozyme C, ... | | Authors: | Tabe, H, Fujita, K, Abe, S, Tsujimoto, M, Kuchimaru, T, Kizaka-Kondo, S, Takano, M, Kitagawa, S, Ueno, T. | | Deposit date: | 2014-08-27 | | Release date: | 2014-12-31 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Preparation of a Cross-Linked Porous Protein Crystal Containing Ru Carbonyl Complexes as a CO-Releasing Extracellular Scaffold
Inorg.Chem., 54, 2015
|
|
4UWV
 
 | | Lysozyme soaked with a ruthenium based CORM with a pyridine ligand (complex 8) | | Descriptor: | CARBON MONOXIDE, CHLORIDE ION, LYSOZYME C, ... | | Authors: | Santos, M.F.A, Mukhopadhyay, A, Romao, M.J, Romao, C.C, Santos-Silva, T. | | Deposit date: | 2014-08-14 | | Release date: | 2014-12-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | A Contribution to the Rational Design of Ru(Co)3Cl2L Complexes for in Vivo Delivery of Co.
Dalton Trans, 44, 2015
|
|
3NNQ
 
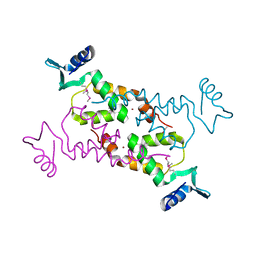 | | Crystal Structure of the N-terminal domain of Moloney murine leukemia virus integrase, Northeast Structural Genomics Consortium Target OR3 | | Descriptor: | ACETATE ION, N-terminal domain of Moloney murine leukemia virus integrase, ZINC ION | | Authors: | Guan, R, Xiao, R, Acton, T, Jiang, M, Roth, M, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-06-24 | | Release date: | 2010-07-14 | | Last modified: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (2.693 Å) | | Cite: | X-ray crystal structure of the N-terminal region of Moloney murine leukemia virus integrase and its implications for viral DNA recognition.
Proteins, 85, 2017
|
|
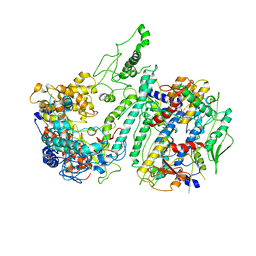
3IKM
 
 | |
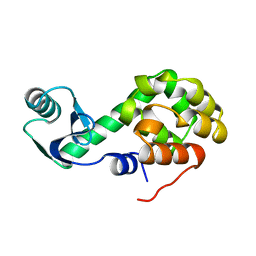
1L47
 
 | |
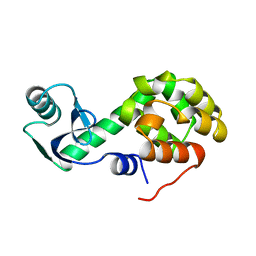
1L60
 
 | |
4UWU
 
 | | Lysozyme soaked with a ruthenium based CORM with a pyridine ligand (complex 7) | | Descriptor: | CARBON MONOXIDE, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Santos, M.F.A, Mukhopadhyay, A, Romao, M.J, Romao, C.C, Santos-Silva, T. | | Deposit date: | 2014-08-14 | | Release date: | 2014-12-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | A Contribution to the Rational Design of Ru(Co)3Cl2L Complexes for in Vivo Delivery of Co.
Dalton Trans, 44, 2015
|
|
1L42
 
 | |
1L46
 
 | |
1L45
 
 | |
3OVE
 
 | |
1L54
 
 | |
1L43
 
 | |
3KIN
 
 | | KINESIN (DIMERIC) FROM RATTUS NORVEGICUS | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, KINESIN HEAVY CHAIN | | Authors: | Kozielski, F, Sack, S, Marx, A, Thormahlen, M, Schonbrunn, E, Biou, V, Thompson, A, Mandelkow, E.-M, Mandelkow, E. | | Deposit date: | 1997-08-25 | | Release date: | 1998-10-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The crystal structure of dimeric kinesin and implications for microtubule-dependent motility.
Cell(Cambridge,Mass.), 91, 1997
|
|
