3N7D
 
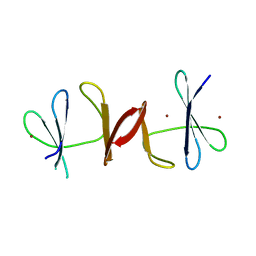 | |
3N1B
 
 | | C-terminal domain of Vps54 subunit of the GARP complex | | Descriptor: | Vacuolar protein sorting-associated protein 54 | | Authors: | Perez-Victoria, F.J, Abascal-Palacios, G, Tascon, I, Kajava, A, Pioro, E.P, Bonifacino, J.S, Hierro, A. | | Deposit date: | 2010-05-15 | | Release date: | 2010-07-14 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.398 Å) | | Cite: | Structural basis for the wobbler mouse neurodegenerative disorder caused by mutation in the Vps54 subunit of the GARP complex.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3NJH
 
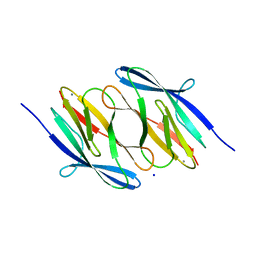 | | D37A mutant of SO1698 protein, an aspartic peptidase from Shewanella oneidensis. | | Descriptor: | CALCIUM ION, GLYCEROL, Peptidase, ... | | Authors: | Osipiuk, J, Mulligan, R, Bargassa, M, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-06-17 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Characterization of member of DUF1888 protein family, self-cleaving and self-assembling endopeptidase.
J.Biol.Chem., 287, 2012
|
|
3NJP
 
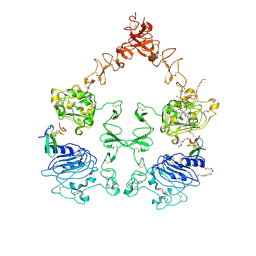 | | The Extracellular and Transmembrane Domain Interfaces in Epidermal Growth Factor Receptor Signaling | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Epidermal growth factor, Epidermal growth factor receptor, ... | | Authors: | Lu, C, Mi, L.-Z, Grey, M.J, Zhu, J, Graef, E, Yokoyama, S, Springer, T.A. | | Deposit date: | 2010-06-17 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.304 Å) | | Cite: | Structural evidence for loose linkage between ligand binding and kinase activation in the epidermal growth factor receptor.
Mol.Cell.Biol., 30, 2010
|
|
3MYQ
 
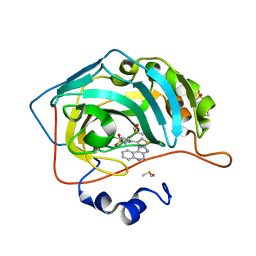 | | Crystal structure of human carbonic anhydrase isozyme II with 2-chloro-5-[(1H-imidazo[4,5-c]quinolin-2-ylsulfanyl)acetyl]benzenesulfonamide | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-chloro-5-[(1H-imidazo[4,5-c]quinolin-2-ylsulfanyl)acetyl]benzenesulfonamide, Carbonic anhydrase 2, ... | | Authors: | Grazulis, S, Manakova, E, Golovenko, D. | | Deposit date: | 2010-05-11 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Indapamide-like benzenesulfonamides as inhibitors of carbonic anhydrases I, II, VII, and XIII.
Bioorg.Med.Chem., 18, 2010
|
|
2ERV
 
 | | Crystal structure of the outer membrane enzyme PagL | | Descriptor: | CALCIUM ION, PENTAETHYLENE GLYCOL MONODECYL ETHER, hypothetical protein Paer03002360 | | Authors: | Rutten, L, Geurtsen, J, Lambert, W, Smolenaers, J.J, Bonvin, A.M, van der Ley, P, Egmond, M.R, Gros, P, Tommassen, J. | | Deposit date: | 2005-10-25 | | Release date: | 2006-04-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure and catalytic mechanism of the LPS 3-O-deacylase PagL from Pseudomonas aeruginosa.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
3N74
 
 | |
3N7U
 
 | | NAD-dependent formate dehydrogenase from higher-plant Arabidopsis thaliana in complex with NAD and azide | | Descriptor: | AZIDE ION, Formate dehydrogenase, GLYCEROL, ... | | Authors: | Shabalin, I.G, Polyakov, K.M, Serov, A.E, Skirgello, O.E, Sadykhov, E.G, Dorovatovskiy, P.V, Tishkov, V.I, Popov, V.O. | | Deposit date: | 2010-05-27 | | Release date: | 2010-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the apo and holo forms of NAD-dependent formate dehydrogenase from the higher-plant Arabidopsis Thaliana
To be Published
|
|
2F0X
 
 | | Crystal structure and function of human thioesterase superfamily member 2(THEM2) | | Descriptor: | SULFATE ION, Thioesterase superfamily member 2 | | Authors: | Cheng, Z, Song, F, Shan, X, Wang, Y, Wei, Z, Gong, W. | | Deposit date: | 2005-11-14 | | Release date: | 2006-10-10 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of human thioesterase superfamily member 2
Biochem.Biophys.Res.Commun., 349, 2006
|
|
3NF4
 
 | |
3NGK
 
 | |
2PJY
 
 | |
3NHB
 
 | | Nucleotide Binding Domain of Human ABCB6 (ADP bound structure) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-binding cassette sub-family B member 6, mitochondrial, ... | | Authors: | Haffke, M, Menzel, A, Carius, Y, Jahn, D, Heinz, D.W. | | Deposit date: | 2010-06-14 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structures of the nucleotide-binding domain of the human ABCB6 transporter and its complexes with nucleotides.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3MXG
 
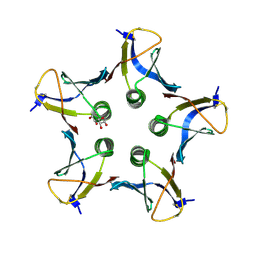 | | Structure of Shiga Toxin type 2 (Stx2) B Pentamer Mutant Q40L | | Descriptor: | D-xylose, Shiga-like toxin 2 subunit B | | Authors: | Kovall, R.A, Vander Wielen, B.D, Friedmann, D.R, Flagler, M.J. | | Deposit date: | 2010-05-07 | | Release date: | 2011-01-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Molecular basis of differential B-pentamer stability of Shiga toxins 1 and 2.
Plos One, 5, 2010
|
|
3N0Z
 
 | |
3N2Y
 
 | |
3N79
 
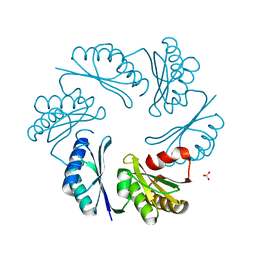 | | PduT C38S Mutant from Salmonella enterica Typhimurium | | Descriptor: | CHLORIDE ION, PduT, SODIUM ION, ... | | Authors: | Crowley, C.S, Cascio, D, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2010-05-26 | | Release date: | 2010-10-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Insight into the Mechanisms of Transport across the Salmonella enterica Pdu Microcompartment Shell.
J.Biol.Chem., 285, 2010
|
|
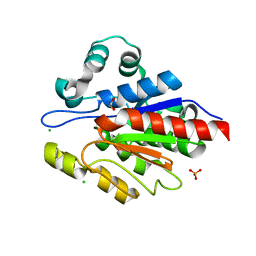
3N7T
 
 | |
3NBR
 
 | | Crystal Structure of Ketosteroid Isomerase D38NP39GD99N from Pseudomonas Testosteroni (tKSI) with 4-Androstene-3,17-dione Bound | | Descriptor: | 4-ANDROSTENE-3-17-DIONE, SULFATE ION, Steroid Delta-isomerase | | Authors: | Gonzalez, A, Tsai, Y, Schwans, J, Ruben, E, Sunden, F, Herschlag, D. | | Deposit date: | 2010-06-03 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal Structure of Ketosteroid Isomerase D38NP39GD99N from Pseudomonas Testosteroni (tKSI) with 4-Androstene-3,17-dione Bound
To be Published
|
|
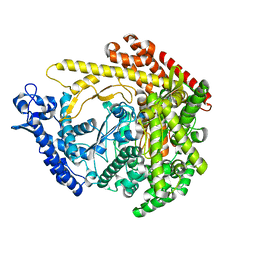
2PO4
 
 | |
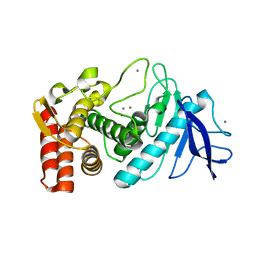
3P7Q
 
 | |
3OWX
 
 | | X-ray Structural study of quinone reductase II inhibition by compounds with micromolar to nanomolar range IC50 values | | Descriptor: | 2-[4-(furan-2-ylcarbonyl)piperazin-1-yl]-6,7-dimethoxyquinazolin-4-amine, FLAVIN-ADENINE DINUCLEOTIDE, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Pegan, S.D, Sturdy, M, Ferry, G, Delagrange, P, Boutin, J.A, Mesecar, A.D. | | Deposit date: | 2010-09-20 | | Release date: | 2011-05-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | X-ray structural studies of quinone reductase 2 nanomolar range inhibitors.
Protein Sci., 20, 2011
|
|
3P8H
 
 | | Crystal structure of L3MBTL1 (MBT repeat) in complex with a nicotinamide antagonist | | Descriptor: | 3-bromo-5-[(4-pyrrolidin-1-ylpiperidin-1-yl)carbonyl]pyridine, GLYCEROL, Lethal(3)malignant brain tumor-like protein, ... | | Authors: | Lam, R, Herold, J.M, Ouyang, H, Tempel, W, Gao, C, Ravichandran, M, Senisterra, G, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Vedadi, M, Kireev, D, Frye, S.V, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-10-13 | | Release date: | 2010-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Small-molecule ligands of methyl-lysine binding proteins.
J.Med.Chem., 54, 2011
|
|
3P91
 
 | |
3OYX
 
 | | Haloferax volcanii Malate Synthase Magnesium/Glyoxylate Complex | | Descriptor: | CHLORIDE ION, GLYOXYLIC ACID, MAGNESIUM ION, ... | | Authors: | Howard, B.R, Bracken, C, Neighbor, A, Thomas, G, Lamlenn, K.K, Schubert, H.L, Whitby, F.G. | | Deposit date: | 2010-09-24 | | Release date: | 2011-06-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal structures of a halophilic archaeal malate synthase from Haloferax volcanii and comparisons with isoforms A and G.
Bmc Struct.Biol., 11, 2011
|
|
