1SS3
 
 | | Solution structure of Ole e 6, an allergen from olive tree pollen | | Descriptor: | Pollen allergen Ole e 6 | | Authors: | Trevino, M.A, Garcia-Mayoral, M.F, Barral, P, Villalba, M, Santoro, J, Rico, M, Rodriguez, R, Bruix, M. | | Deposit date: | 2004-03-23 | | Release date: | 2004-08-03 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of Ole e 6, a Major Allergen from Olive Tree Pollen.
J.Biol.Chem., 279, 2004
|
|
6ALK
 
 | | NMR solution structure of the major beech pollen allergen Fag s 1 | | Descriptor: | Fag s 1 pollen allergen | | Authors: | Moraes, A.H, Asam, A, Almeida, F.C.L, Wallner, M, Ferreira, F, Valente, A.P. | | Deposit date: | 2017-08-08 | | Release date: | 2018-08-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for cross-reactivity and conformation fluctuation of the major beech pollen allergen Fag s 1.
Sci Rep, 8, 2018
|
|
6SL5
 
 | | Dunaliella Photosystem I Supercomplex | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | Authors: | Nelson, N, Caspy, I, Malavath, T, Klaiman, D, Shkolinsky, Y. | | Deposit date: | 2019-08-18 | | Release date: | 2020-06-24 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Structure and energy transfer pathways of the Dunaliella Salina photosystem I supercomplex.
Biochim Biophys Acta Bioenerg, 1861, 2020
|
|
5MMU
 
 | | NMR solution structure of the major apple allergen Mal d 1 | | Descriptor: | Major allergen Mal d 1 | | Authors: | Ahammer, L, Grutsch, S, Kamenik, A.S, Liedl, K.R, Tollinger, M. | | Deposit date: | 2016-12-12 | | Release date: | 2017-02-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the Major Apple Allergen Mal d 1.
J. Agric. Food Chem., 65, 2017
|
|
1PP4
 
 | |
8DB4
 
 | |
1W2U
 
 | | X-RAY CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF HUMICOLA GRISEA CEL12A IN COMPLEX WITH A SOAKED THIO CELLOTETRAOSE | | Descriptor: | ENDOGLUCANASE, SULFATE ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Berglund, G.I, Shaw, A, Stahlberg, J, Kenne, L, Driguez, T.H, Mitchinson, C, Sandgren, M. | | Deposit date: | 2004-07-08 | | Release date: | 2004-09-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystal Complex Structures Reveal How Substrate is Bound in the -4 to the +2 Binding Sites of Humicola Grisea Cel12A
J.Mol.Biol., 342, 2004
|
|
1UU6
 
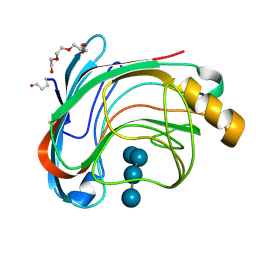 | | X-RAY CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF HUMICOLA GRISEA CEL12A IN COMPLEX WITH A SOAKED CELLOPENTAOSE | | Descriptor: | ENDO-BETA-1,4-GLUCANASE, TETRAETHYLENE GLYCOL, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Berglund, G.I, Shaw, A, Stahlberg, J, Kenne, L, Driguez, T.H, Mitchinson, C, Sandgren, M. | | Deposit date: | 2003-12-15 | | Release date: | 2004-09-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Complex Structures Reveal How Substrate is Bound in the -4 to the +2 Binding Sites of Humicola Grisea Cel12A
J.Mol.Biol., 342, 2004
|
|
1UU4
 
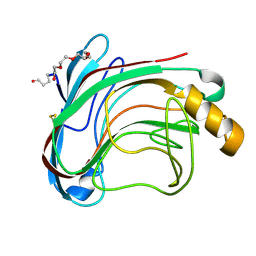 | | X-RAY CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF HUMICOLA GRISEA CEL12A IN COMPLEX WITH CELLOBIOSE | | Descriptor: | ENDO-BETA-1,4-GLUCANASE, TETRAETHYLENE GLYCOL, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Berglund, G.I, Shaw, A, Stahlberg, J, Kenne, L, Driguez, T.H, Mitchinson, C, Sandgren, M. | | Deposit date: | 2003-12-15 | | Release date: | 2004-09-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Crystal Complex Structures Reveal How Substrate is Bound in the -4 to the +2 Binding Sites of Humicola Grisea Cel12A
J.Mol.Biol., 342, 2004
|
|
1UU5
 
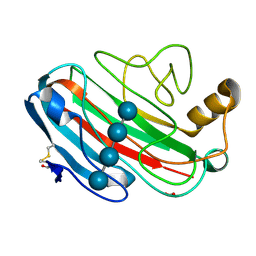 | | X-RAY CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF HUMICOLA GRISEA CEL12A SOAKED WITH CELLOTETRAOSE | | Descriptor: | ACETATE ION, ENDO-BETA-1,4-GLUCANASE, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Berglund, G.I, Shaw, A, Stahlberg, J, Kenne, L, Driguez, T.H, Mitchinson, C, Sandgren, M. | | Deposit date: | 2003-12-15 | | Release date: | 2004-09-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Crystal Complex Structures Reveal How Substrate is Bound in the -4 to the +2 Binding Sites of Humicola Grisea Cel12A
J.Mol.Biol., 342, 2004
|
|
7ZRI
 
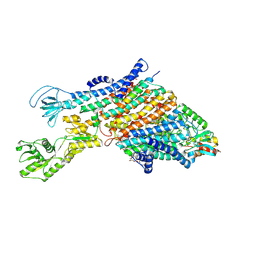 | | Cryo-EM structure of the KdpFABC complex in a nucleotide-free E1 conformation loaded with K+ | | Descriptor: | CARDIOLIPIN, POTASSIUM ION, Potassium-transporting ATPase ATP-binding subunit, ... | | Authors: | Hielkema, L, Stock, C, Silberberg, J.M, Corey, R.A, Wunnicke, D, Stansfeld, P.J, Haenelt, I, Paulino, C. | | Deposit date: | 2022-05-04 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Inhibited KdpFABC transitions into an E1 off-cycle state.
Elife, 11, 2022
|
|
4BK7
 
 | |
6X8P
 
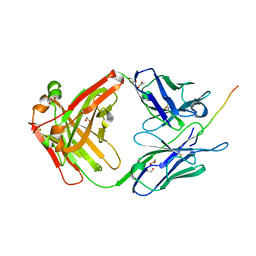 | |
6Q3R
 
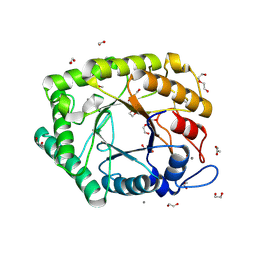 | | ASPERGILLUS ACULEATUS GALACTANASE | | Descriptor: | 1,2-ETHANEDIOL, 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, ACETATE ION, ... | | Authors: | Muderspach, S.J, Torpenholt, S, Lo Leggio, L, Poulsen, J.C.N. | | Deposit date: | 2018-12-04 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structure of Aspergillus aculeatus beta-1,4-galactanase in complex with galactobiose.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
2JMH
 
 | | NMR solution structure of Blo t 5, a major mite allergen from Blomia tropicalis | | Descriptor: | Mite allergen Blo t 5 | | Authors: | Naik, M.T, Chang, C, Kuo, I, Chua, K, Huang, T. | | Deposit date: | 2006-11-12 | | Release date: | 2007-11-13 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Roles of Structure and Structural Dynamics in the Antibody Recognition of the Allergen Proteins: An NMR Study on Blomia tropicalis Major Allergen
Structure, 16, 2008
|
|
7ZRG
 
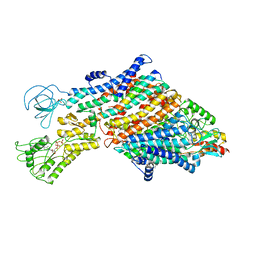 | | Cryo-EM map of the WT KdpFABC complex in the E1_ATPearly conformation, under turnover conditions | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CARDIOLIPIN, POTASSIUM ION, ... | | Authors: | Hielkema, L, Stock, C, Silberberg, J.M, Corey, R.A, Rheinberger, J, Wunnicke, D, Dubach, V.R.A, Stansfeld, P.J, Haenelt, I, Paulino, C. | | Deposit date: | 2022-05-04 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Inhibited KdpFABC transitions into an E1 off-cycle state.
Elife, 11, 2022
|
|
5KSB
 
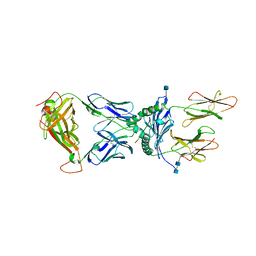 | | T15-DQ8.5-glia-gamma1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DQ8.5-glia-gamma1 peptide, ... | | Authors: | Petersen, J, Rossjohn, J, Reid, H.H. | | Deposit date: | 2016-07-08 | | Release date: | 2016-09-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Diverse T Cell Receptor Gene Usage in HLA-DQ8-Associated Celiac Disease Converges into a Consensus Binding Solution.
Structure, 24, 2016
|
|
5KS9
 
 | | Bel502-DQ8-glia-alpha1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Bel502 TCR alpha TRAV20*01, Bel502 TCR beta TRBV9*01, ... | | Authors: | Petersen, J, Rossjohn, J, Reid, H.H. | | Deposit date: | 2016-07-08 | | Release date: | 2016-09-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Diverse T Cell Receptor Gene Usage in HLA-DQ8-Associated Celiac Disease Converges into a Consensus Binding Solution.
Structure, 24, 2016
|
|
7UUR
 
 | | The 1.67 Angstrom CryoEM structure of the [NiFe]-hydrogenase Huc from Mycobacterium smegmatis - catalytic dimer (Huc2S2L) | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, HYDROXIDE ION, ... | | Authors: | Grinter, R, Venugopal, H, Kropp, A, Greening, C. | | Deposit date: | 2022-04-28 | | Release date: | 2023-01-04 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (1.67 Å) | | Cite: | Structural basis for bacterial energy extraction from atmospheric hydrogen.
Nature, 615, 2023
|
|
7UUS
 
 | | The CryoEM structure of the [NiFe]-hydrogenase Huc from Mycobacterium smegmatis - Full complex focused refinement of stalk | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, Hydrogenase-2, ... | | Authors: | Grinter, R, Venugopal, H, Kropp, A, Greening, C. | | Deposit date: | 2022-04-28 | | Release date: | 2023-01-04 | | Last modified: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Structural basis for bacterial energy extraction from atmospheric hydrogen.
Nature, 615, 2023
|
|
7UTD
 
 | | The 2.19-angstrom CryoEM structure of the [NiFe]-hydrogenase Huc from Mycobacterium smegmatis - Complex minus stalk | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, Hydrogenase-2, ... | | Authors: | Grinter, R, Venugopal, H, Kropp, A, Greening, C. | | Deposit date: | 2022-04-26 | | Release date: | 2023-01-04 | | Last modified: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (2.19 Å) | | Cite: | Structural basis for bacterial energy extraction from atmospheric hydrogen.
Nature, 615, 2023
|
|
7ZRM
 
 | | Cryo-EM map of the unphosphorylated KdpFABC complex in the E1-P_ADP conformation, under turnover conditions | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Hielkema, L, Stock, C, Silberberg, J.M, Corey, R.A, Wunnicke, D, Dubach, V.R.A, Stansfeld, P.J, Haenelt, I, Paulino, C. | | Deposit date: | 2022-05-04 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Inhibited KdpFABC transitions into an E1 off-cycle state.
Elife, 11, 2022
|
|
7ZRD
 
 | | Cryo-EM map of the WT KdpFABC complex in the E1-P tight conformation, stabilised with the inhibitor orthovanadate | | Descriptor: | CARDIOLIPIN, POTASSIUM ION, Potassium-transporting ATPase ATP-binding subunit, ... | | Authors: | Hielkema, L, Stock, C, Silberberg, J.M, Corey, R.A, Wunnicke, D, Stansfeld, P.J, Haenelt, I, Paulino, C. | | Deposit date: | 2022-05-04 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Inhibited KdpFABC transitions into an E1 off-cycle state.
Elife, 11, 2022
|
|
7ZRL
 
 | | Cryo-EM map of the unphosphorylated KdpFABC complex in the E2-P conformation, under turnover conditions | | Descriptor: | POTASSIUM ION, Potassium-transporting ATPase ATP-binding subunit, Potassium-transporting ATPase KdpC subunit, ... | | Authors: | Hielkema, L, Stock, C, Silberberg, J.M, Corey, R.A, Wunnicke, D, Dubach, V.R.A, Stansfeld, P.J, Haenelt, I, Paulino, C. | | Deposit date: | 2022-05-04 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Inhibited KdpFABC transitions into an E1 off-cycle state.
Elife, 11, 2022
|
|
7ZRE
 
 | | Cryo-EM map of the WT KdpFABC complex in the E1-P tight conformation, under turnover conditions | | Descriptor: | CARDIOLIPIN, POTASSIUM ION, Potassium-transporting ATPase ATP-binding subunit, ... | | Authors: | Hielkema, L, Stock, C, Silberberg, J.M, Corey, R.A, Wunnicke, D, Dubach, V.R.A, Stansfeld, P.J, Haenelt, I, Paulino, C. | | Deposit date: | 2022-05-04 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Inhibited KdpFABC transitions into an E1 off-cycle state.
Elife, 11, 2022
|
|
