8R35
 
 | |
8R34
 
 | | CryoEM structure of the symmetric Pho90 dimer from yeast with substrates. | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, Low-affinity phosphate transporter PHO90, PHOSPHATE ION, ... | | Authors: | Schneider, S, Kuehlbrandt, W, Yildiz, O. | | Deposit date: | 2023-11-08 | | Release date: | 2024-04-24 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Complementary structures of the yeast phosphate transporter Pho90 provide insights into its transport mechanism.
Structure, 32, 2024
|
|
8R8T
 
 | |
3HZZ
 
 | |
8QTA
 
 | | Cryo-EM structure of Streptococcus pneumoniae NADPH oxidase F397A mutant in complex with NADPH | | Descriptor: | FAD-binding FR-type domain-containing protein, FLAVIN-ADENINE DINUCLEOTIDE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Dubach, V.R.A, San Segundo-Acosta, P, Murphy, B.J. | | Deposit date: | 2023-10-12 | | Release date: | 2024-07-24 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Structural and mechanistic insights into Streptococcus pneumoniae NADPH oxidase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
5C20
 
 | | Crystal structure of EV71 3C Proteinase in complex with Compound 2 | | Descriptor: | 2-methylpropyl N-[(2S)-1-oxidanylidene-1-[[(2S)-1-oxidanyl-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-3-phenyl-propan-2-yl]carbamate, 3C proteinase | | Authors: | Zhang, L, Huang, G, Cai, Q, Zhao, C, Ren, H, Li, P, Li, N, Chen, S, Li, J, Lin, T. | | Deposit date: | 2015-06-15 | | Release date: | 2016-06-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Optimize the interactions at S4 with efficient inhibitors targeting 3C proteinase from enterovirus 71
J.Mol.Recognit., 29, 2016
|
|
8QT6
 
 | |
8QT9
 
 | | Cryo-EM structure of stably reduced Streptococcus pneumoniae NADPH oxidase in complex with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, FAD-binding FR-type domain-containing protein, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Dubach, V.R.A, San Segundo-Acosta, P, Murphy, B.J. | | Deposit date: | 2023-10-12 | | Release date: | 2024-07-24 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (2.36 Å) | | Cite: | Structural and mechanistic insights into Streptococcus pneumoniae NADPH oxidase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8QZP
 
 | | Structure of the non-mitochondrial citrate synthase from Ananas comosus | | Descriptor: | Citrate synthase | | Authors: | Lo, Y.K, Bohn, S, Sendker, F.L, Schuller, J.M, Hochberg, G. | | Deposit date: | 2023-10-28 | | Release date: | 2024-07-24 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (4.15 Å) | | Cite: | Frequent transitions in self-assembly across the evolution of a central metabolic enzyme.
Biorxiv, 2024
|
|
8QT7
 
 | | Cryo-EM structure of Streptococcus pneumoniae NADPH oxidase in complex with NADPH | | Descriptor: | FAD-binding FR-type domain-containing protein, FLAVIN-ADENINE DINUCLEOTIDE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Dubach, V.R.A, San Segundo-Acosta, P, Murphy, B.J. | | Deposit date: | 2023-10-12 | | Release date: | 2024-07-24 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Structural and mechanistic insights into Streptococcus pneumoniae NADPH oxidase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
4ZRZ
 
 | | PlyCB mutant R66E | | Descriptor: | PlyCB | | Authors: | Gallagher, D.T, Nelson, D.C, Shen, Y. | | Deposit date: | 2015-05-12 | | Release date: | 2016-09-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | A bacteriophage endolysin that eliminates intracellular streptococci.
Elife, 5, 2016
|
|
4ZZ7
 
 | | Crystal structure of methylmalonate-semialdehyde dehydrogenase (DddC) from Oceanimonas doudoroffii | | Descriptor: | Methylmalonate-semialdehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Do, H, Lee, C.W, Lee, S.G, Kang, H, Park, C.M, Kim, H.J, Park, H, Park, H, Lee, J.H. | | Deposit date: | 2015-05-22 | | Release date: | 2016-04-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure and modeling of the tetrahedral intermediate state of methylmalonate-semialdehyde dehydrogenase (MMSDH) from Oceanimonas doudoroffii.
J. Microbiol., 54, 2016
|
|
3HO7
 
 | |
5A2B
 
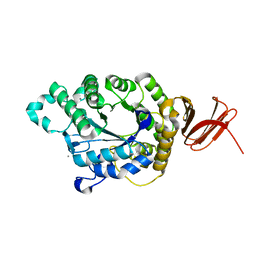 | | Crystal Structure of Anoxybacillus Alpha-amylase Provides Insights into a New Glycosyl Hydrolase Subclass | | Descriptor: | ANOXYBACILLUS ALPHA-AMYLASE, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Ng, C.L, Chai, K.P, Othman, N.F, Teh, A.H, Ho, K.L, Chan, K.G, Goh, K.M. | | Deposit date: | 2015-05-17 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of Anoxybacillus Alpha-Amylase Provides Insights Into Maltose Binding of a New Glycosyl Hydrolase Subclass.
Sci.Rep., 6, 2016
|
|
5C1E
 
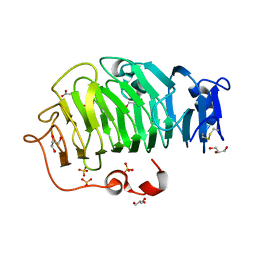 | | Crystal Structure of the Pectin Methylesterase from Aspergillus niger in Penultimately Deglycosylated Form (N-acetylglucosamine Stub at Asn84) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, CHLORIDE ION, ... | | Authors: | Jameson, G.B, Williams, M.A.K, Loo, T.S, Kent, L.M, Melton, L.D, Mercadante, D. | | Deposit date: | 2015-06-13 | | Release date: | 2015-07-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and Properties of a Non-processive, Salt-requiring, and Acidophilic Pectin Methylesterase from Aspergillus niger Provide Insights into the Key Determinants of Processivity Control.
J.Biol.Chem., 291, 2016
|
|
8QPL
 
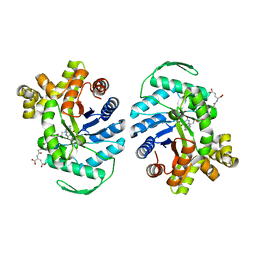 | | F420-Dependent Methylene-Tetrahydromethanopterin Reductase with F420 from Methanocaldococcus jannaschii | | Descriptor: | 5,10-methylenetetrahydromethanopterin reductase, COENZYME F420 | | Authors: | Gehl, M, Demmer, U, Ermler, U, Shima, S. | | Deposit date: | 2023-10-02 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mutational and structural studies of ( beta alpha ) 8 -barrel fold methylene-tetrahydropterin reductases utilizing a common catalytic mechanism.
Protein Sci., 33, 2024
|
|
3G29
 
 | |
8QPJ
 
 | |
3G1I
 
 | |
8QPM
 
 | | Structure of methylene-tetrahydromethanopterin reductase from Methanocaldococcus jannaschii | | Descriptor: | 5,10-methylenetetrahydromethanopterin reductase | | Authors: | Gehl, M, Demmer, U, Ermler, U, Shima, S. | | Deposit date: | 2023-10-02 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mutational and structural studies of ( beta alpha ) 8 -barrel fold methylene-tetrahydropterin reductases utilizing a common catalytic mechanism.
Protein Sci., 33, 2024
|
|
8QQ8
 
 | | Crystal Structure of F420-dependent Methylene-Tetrahydromethanopterin Reductase Mutant E6Q from Methanocaldococcus Jannaschii | | Descriptor: | 5,10-methylenetetrahydromethanopterin reductase | | Authors: | Gehl, M, Demmer, U, Ermler, U, Shima, S. | | Deposit date: | 2023-10-04 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutational and structural studies of ( beta alpha ) 8 -barrel fold methylene-tetrahydropterin reductases utilizing a common catalytic mechanism.
Protein Sci., 33, 2024
|
|
8QMW
 
 | | Non-obligately L8S8-complex forming RubisCO derived from ancestral sequence reconstruction and rational engineering in L8S8 complex with substitutions R269W, E271R, L273N | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, ... | | Authors: | Zarzycki, J, Schulz, L, Erb, T.J, Hochberg, G.K.A. | | Deposit date: | 2023-09-25 | | Release date: | 2024-10-02 | | Last modified: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Layered entrenchment maintains essentiality in the evolution of Form I Rubisco complexes.
Embo J., 44, 2025
|
|
3TFB
 
 | | Transthyretin natural mutant A25T | | Descriptor: | Transthyretin | | Authors: | Azevedo, E.P.C, Pereira, H.M, Garratt, R.C, Kelly, J.W, Foguel, D, Palhano, F.L. | | Deposit date: | 2011-08-15 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.033 Å) | | Cite: | Dissecting the Structure, Thermodynamic Stability, and Aggregation Properties of the A25T Transthyretin (A25T-TTR) Variant Involved in Leptomeningeal Amyloidosis: Identifying Protein Partners That Co-Aggregate during A25T-TTR Fibrillogenesis in Cerebrospinal Fluid.
Biochemistry, 50, 2011
|
|
8QMQ
 
 | | Succinic semialdehyde dehydrogenase from E. coli with bound NAD+ | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Succinate semialdehyde dehydrogenase [NAD(P)+] Sad | | Authors: | He, H, Zarzycki, J, Erb, T.J. | | Deposit date: | 2023-09-25 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Adaptive laboratory evolution recruits the promiscuity of succinate semialdehyde dehydrogenase to repair different metabolic deficiencies.
Nat Commun, 15, 2024
|
|
8QMV
 
 | | L2 forming RubisCO derived from ancestral sequence reconstruction of the last common ancestor of Form I'' and Form I RubisCOs | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, RubisCO large subunit | | Authors: | Zarzycki, J, Schulz, L, Erb, T.J, Hochberg, G.K.A. | | Deposit date: | 2023-09-25 | | Release date: | 2024-10-16 | | Last modified: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Layered entrenchment maintains essentiality in the evolution of Form I Rubisco complexes.
Embo J., 44, 2025
|
|
