9G8J
 
 | |
9GCA
 
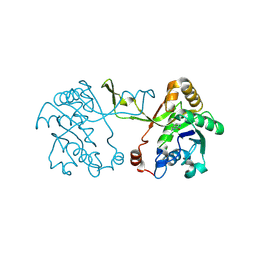 | |
1MNZ
 
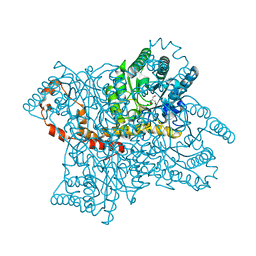 | | Atomic structure of Glucose isomerase | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Nowak, E, Panjikar, S, Tucker, P.A. | | Deposit date: | 2002-09-06 | | Release date: | 2002-09-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | Atomic structure of Glucose isomerase
To be published
|
|
9GSU
 
 | |
1MOJ
 
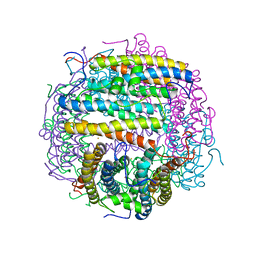 | | Crystal structure of an archaeal dps-homologue from Halobacterium salinarum | | Descriptor: | Dps-like ferritin, FE (III) ION, MAGNESIUM ION, ... | | Authors: | Zeth, K, Offermann, S, Essen, L.O, Oesterhelt, D. | | Deposit date: | 2002-09-09 | | Release date: | 2004-04-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Iron-oxo clusters biomineralizing on protein surfaces: Structural analysis of Halobacterium salinarum DpsA in its low- and high-iron states.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
9GA6
 
 | | The crystal structure of human Annexin A4 derived from crystals grown in 40 mM of CaCl2 | | Descriptor: | Annexin A4, CALCIUM ION, GLYCEROL, ... | | Authors: | Vitagliano, L, Barra, G, Ghilardi, O, Di Micco, S, Bifulco, G, Campiglia, P, Sala, M, Scala, M.C, Ruggiero, A. | | Deposit date: | 2024-07-26 | | Release date: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Computational, crystallographic, and biophysical characterizations provide insights into calcium and phosphate binding by human annexin A4.
Int.J.Biol.Macromol., 308, 2025
|
|
1MOY
 
 | | Streptavidin Mutant with Osteopontin Hexapeptide Insertion Including RGD | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Streptavidin | | Authors: | Le Trong, I, McDevitt, T.C, Nelson, K.E, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 2002-09-10 | | Release date: | 2003-05-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural characterization and comparison of RGD cell-adhesion recognition sites engineered into streptavidin.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
9G4M
 
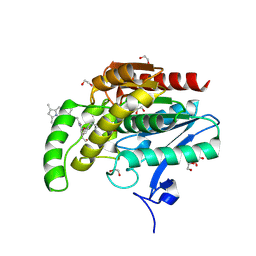 | | Crystal structure of monoacylglycerol lipase with BODIPY labeled probe | | Descriptor: | 1,2-ETHANEDIOL, 3-[2,2-bis(fluoranyl)-10,12-dimethyl-1$l^{4},3-diaza-2$l^{4}-boratricyclo[7.3.0.0^{3,7}]dodeca-1(12),4,6,8,10-pentaen-4-yl]-~{N}-[2-[2-[[2-[(6-oxidanylidene-7-oxa-2,5-diazaspiro[3.4]octan-2-yl)carbonyl]-2-azaspiro[3.3]heptan-6-yl]methyl]phenoxy]ethyl]propanamide, Monoglyceride lipase | | Authors: | Guberman, M, Hentsch, A, Radetzki, S, Kaushik, S, Huizenga, M, van der Stelt, M, Paul, J, Schippers, M, Hochstrasser, R, von Kries, J.P, Blaising, J, Lipstein, N, Nazare, M, Grether, U, Kuhn, B, Walter, A, Benz, J. | | Deposit date: | 2024-07-15 | | Release date: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | A Highly Selective and Versatile Probe Platform for Visualization of Monoacylglycerol Lipase.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
9GTS
 
 | | Cryo-EM structure of a contractile injection system in Streptomyces coelicolor, the cap portion in extended state. | | Descriptor: | Phage tail protein, Phage tail sheath family protein, Pvc16 N-terminal domain-containing protein | | Authors: | Casu, B, Sallmen, J.W, Hass, P.E, Afanasyev, P, Xu, J, Schlimpert, S, Pilhofer, M. | | Deposit date: | 2024-09-18 | | Release date: | 2025-06-18 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Function and firing of the Streptomyces coelicolor contractile injection system requires the membrane protein CisA.
Elife, 14, 2025
|
|
9G28
 
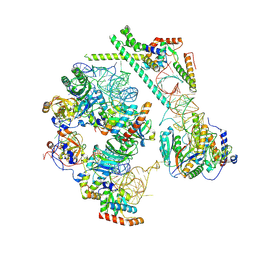 | | snR30 snoRNP - State 2 - Utp23-Krr1-deltaC3 | | Descriptor: | 40S ribosomal protein S13, 40S ribosomal protein S14-A, H/ACA ribonucleoprotein complex subunit CBF5, ... | | Authors: | Thoms, M, Berninghausen, O, Beckmann, R. | | Deposit date: | 2024-07-10 | | Release date: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | H/ACA snR30 snoRNP guides independent 18S rRNA subdomain formation.
Nat Commun, 16, 2025
|
|
1MI1
 
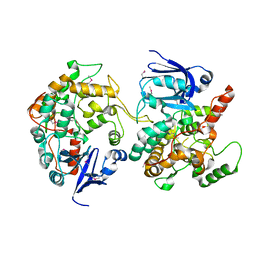 | | Crystal Structure of the PH-BEACH Domain of Human Neurobeachin | | Descriptor: | Neurobeachin | | Authors: | Jogl, G, Shen, Y, Gebauer, D, Li, J, Wiegmann, K, Kashkar, H, Kroenke, M, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-08-21 | | Release date: | 2002-09-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the BEACH domain reveals an unusual fold and extensive association with a novel PH domain.
EMBO J., 21, 2002
|
|
9G1V
 
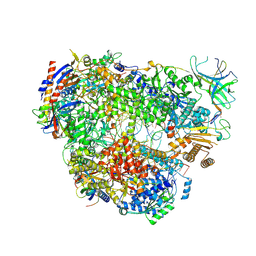 | | Yeast RNA polymerase I elongation complex stalled by an apurinic site | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Santos-Aledo, A, Plaza-Pegueroles, A, Ruiz, F.M, Fernandez-Tornero, C. | | Deposit date: | 2024-07-10 | | Release date: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM uncovers a sequential mechanism for RNA polymerase I pausing and stalling at abasic DNA lesions.
Nat Commun, 16, 2025
|
|
9G25
 
 | | snR30 snoRNP - State 1 - Utp23-Krr1-deltaC3 | | Descriptor: | 40S ribosomal protein S13, 40S ribosomal protein S14-A, H/ACA ribonucleoprotein complex subunit CBF5, ... | | Authors: | Thoms, M, Berninghausen, O, Beckmann, R. | | Deposit date: | 2024-07-10 | | Release date: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | H/ACA snR30 snoRNP guides independent 18S rRNA subdomain formation.
Nat Commun, 16, 2025
|
|
1MIQ
 
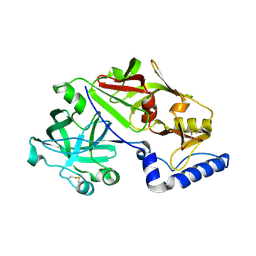 | | Crystal structure of proplasmepsin from the human malarial pathogen Plasmodium vivax | | Descriptor: | plasmepsin | | Authors: | Bernstein, N.K, Cherney, M.M, Yowell, C.A, Dame, J.B, James, M.N. | | Deposit date: | 2002-08-23 | | Release date: | 2002-09-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the activation of P. vivax plasmepsin.
J.Mol.Biol., 329, 2003
|
|
1MP8
 
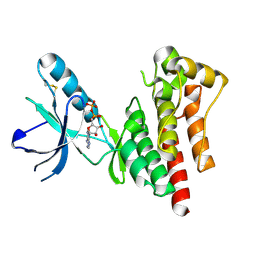 | | Crystal structure of Focal Adhesion Kinase (FAK) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, focal adhesion kinase 1 | | Authors: | Nowakowski, J, Cronin, C.N, McRee, D.E, Knuth, M.W, Nelson, C.G, Pavletich, N.P, Rodgers, J, Sang, B.-C, Scheibe, D.N, Swanson, R.V, Thompson, D.A. | | Deposit date: | 2002-09-11 | | Release date: | 2003-09-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of the cancer-related Aurora-A, FAK, and EphA2 protein kinases from nanovolume crystallography
Structure, 10, 2002
|
|
9G26
 
 | | Yeast RNA polymerase I elongation complex stalled by an apurinic site, closed state | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Santos-Aledo, A, Plaza-Pegueroles, A, Ruiz, F.M, Fernandez-Tornero, C. | | Deposit date: | 2024-07-10 | | Release date: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM uncovers a sequential mechanism for RNA polymerase I pausing and stalling at abasic DNA lesions.
Nat Commun, 16, 2025
|
|
9G5M
 
 | |
1MPR
 
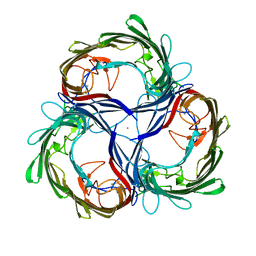 | | MALTOPORIN FROM SALMONELLA TYPHIMURIUM | | Descriptor: | CALCIUM ION, MALTOPORIN | | Authors: | Meyer, J.E.W, Schulz, G.E. | | Deposit date: | 1996-12-18 | | Release date: | 1997-03-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of maltoporin from Salmonella typhimurium ligated with a nitrophenyl-maltotrioside.
J.Mol.Biol., 266, 1997
|
|
9GFY
 
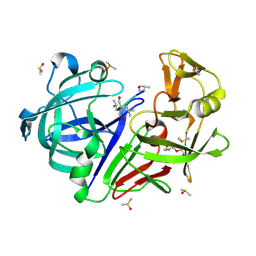 | | Endothiapepsin in complex with pepstatin soaked at pH 7.6 | | Descriptor: | DIMETHYL SULFOXIDE, Endothiapepsin, PENTAETHYLENE GLYCOL, ... | | Authors: | Mueller, J.M, Glinca, S. | | Deposit date: | 2024-08-12 | | Release date: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Protonation Effects in Protein-Ligand Complexes - A Case Study of Endothiapepsin and Pepstatin A with Computational and Experimental Methods.
Chemmedchem, 20, 2025
|
|
7RL3
 
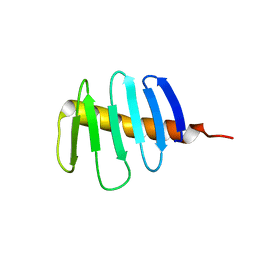 | | Structure of Drosophila melanogaster Plk4 PB3 | | Descriptor: | Serine/threonine-protein kinase PLK4 | | Authors: | Slep, K.C, Slevin, L.K. | | Deposit date: | 2021-07-23 | | Release date: | 2022-08-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Polo-like kinase 4 homodimerization and condensate formation regulate its own protein levels but are not required for centriole assembly.
Mol.Biol.Cell, 34, 2023
|
|
1MJ9
 
 | | Crystal structure of yeast Esa1(C304S) mutant complexed with Coenzyme A | | Descriptor: | COENZYME A, ESA1 PROTEIN, SODIUM ION | | Authors: | Yan, Y, Harper, S, Speicher, D, Marmorstein, R. | | Deposit date: | 2002-08-27 | | Release date: | 2002-10-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The catalytic mechanism of the ESA1 histone acetyltransferase involves a self-acetylated intermediate.
Nat.Struct.Biol., 9, 2002
|
|
9GTR
 
 | | Cryo-EM structure of a contractile injection system in Streptomyces coelicolor, the baseplate complex in extended state applied 3-fold symmetry. | | Descriptor: | Gp5/Type VI secretion system Vgr protein OB-fold domain-containing protein | | Authors: | Casu, B, Sallmen, J.W, Hass, P.E, Afanasyev, P, Xu, J, Schlimpert, S, Pilhofer, M. | | Deposit date: | 2024-09-18 | | Release date: | 2025-06-18 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Function and firing of the Streptomyces coelicolor contractile injection system requires the membrane protein CisA.
Elife, 14, 2025
|
|
1MJQ
 
 | |
9GAD
 
 | | Structure and catalytic mechanism of SAM-AMP lyase in Clostridium botulinum CorA-associated type III CRISPR system | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, SAM-AMP Lyase | | Authors: | McMahon, S.A, Gloster, T.M, White, M.F, Graham, S, Chi, H. | | Deposit date: | 2024-07-26 | | Release date: | 2025-07-02 | | Last modified: | 2025-07-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | SAM-AMP lyases in type III CRISPR defence.
Nucleic Acids Res., 53, 2025
|
|
9G5H
 
 | | p53-Y220C Core Domain Covalently Bound to 2-chloro-5-cyanopyrazine Soaked at 5 mM | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5-chloranylpyrazine-2-carbonitrile, ... | | Authors: | Stahlecker, J, Klett, T, Stehle, T, Boeckler, F.M. | | Deposit date: | 2024-07-17 | | Release date: | 2025-06-11 | | Last modified: | 2025-07-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | S N Ar Reactive Pyrazine Derivatives as p53-Y220C Cleft Binders with Diverse Binding Modes.
Drug Des Devel Ther, 19, 2025
|
|
