1MNM
 
 | |
9GNZ
 
 | | Salmonella cap-filament complex | | Descriptor: | Flagellar hook-associated protein 2, Flagellin | | Authors: | Qin, K, Einenkel, R, Erhardt, M, Bergeron, J.R.C. | | Deposit date: | 2024-09-04 | | Release date: | 2025-04-09 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The structure of the complete extracellular bacterial flagellum reveals the mechanism of flagellin incorporation.
Nat Microbiol, 10, 2025
|
|
7RV9
 
 | | Crystal structure of the BCL6 BTB domain in complex with OICR-10269 | | Descriptor: | DIMETHYL SULFOXIDE, Isoform 2 of B-cell lymphoma 6 protein, N-[5-chloro-2-(4-methylpiperazin-1-yl)pyridin-4-yl]-2-{5-(3-cyano-4-hydroxy-5-methylphenyl)-3-[3-(1-methyl-1H-pyrazol-4-yl)prop-2-yn-1-yl]-4-oxo-3,4-dihydro-7H-pyrrolo[2,3-d]pyrimidin-7-yl}acetamide | | Authors: | Kuntz, D.A, Prive, G.G. | | Deposit date: | 2021-08-18 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the BCL6 BTB domain
To Be Published
|
|
9GH4
 
 | | Fusobacterium nucleatum adhesin CbpF | | Descriptor: | Cell surface protein | | Authors: | Roderer, D, Marongiu, G.L, Fink, U. | | Deposit date: | 2024-08-15 | | Release date: | 2025-04-02 | | Last modified: | 2025-04-23 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for immune cell binding of Fusobacterium nucleatum via the trimeric autotransporter adhesin CbpF.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
1MGN
 
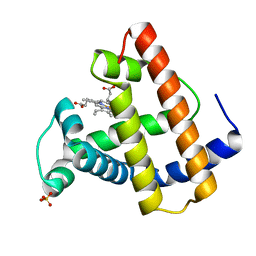 | |
9G0F
 
 | | CryoEM structure of PmcTnsC-dsDNA-AMPPNP | | Descriptor: | AAA+ ATPase domain-containing protein, DNA, MAGNESIUM ION, ... | | Authors: | Finocchio, G, Chanez, C, Querques, I, Speichert, K.J, Jinek, M. | | Deposit date: | 2024-07-08 | | Release date: | 2025-04-02 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of TnsC oligomerization and transposase recruitment in type I-B CRISPR-associated transposons.
Nucleic Acids Res., 53, 2025
|
|
7RVJ
 
 | |
1MJZ
 
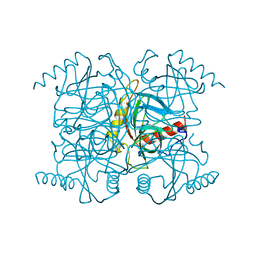 | | STRUCTURE OF INORGANIC PYROPHOSPHATASE MUTANT D97N | | Descriptor: | INORGANIC PYROPHOSPHATASE | | Authors: | Oganesyan, V, Harutyunyan, E.H, Avaeva, S.M, Huber, R. | | Deposit date: | 1997-02-08 | | Release date: | 1997-12-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-dimensional structures of mutant forms of E. coli inorganic pyrophosphatase with Asp-->Asn single substitution in positions 42, 65, 70, and 97.
Biochemistry Mosc., 63, 1998
|
|
1MII
 
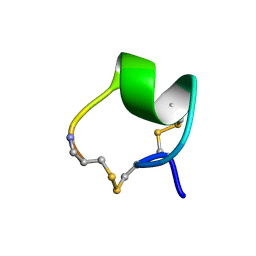 | | SOLUTION STRUCTURE OF ALPHA-CONOTOXIN MII | | Descriptor: | PROTEIN (ALPHA CONOTOXIN MII) | | Authors: | Hill, J.M, Oomen, C.J, Miranda, L.P, Bingham, J.P, Alewood, P.F, Craik, D.J. | | Deposit date: | 1998-10-05 | | Release date: | 1998-10-21 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of alpha-conotoxin MII by NMR spectroscopy: effects of solution environment on helicity.
Biochemistry, 37, 1998
|
|
1MIR
 
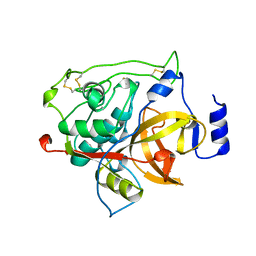 | | RAT PROCATHEPSIN B | | Descriptor: | PROCATHEPSIN B | | Authors: | Cygler, M, Sivaraman, J, Grochulski, P, Coulombe, R, Storer, A.C, Mort, J.S. | | Deposit date: | 1996-01-12 | | Release date: | 1997-01-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of rat procathepsin B: model for inhibition of cysteine protease activity by the proregion.
Structure, 4, 1996
|
|
7RRD
 
 | | IDO1 IN COMPLEX WITH COMPOUND S-1 | | Descriptor: | 3-[4-(1H-benzimidazol-2-yl)phenyl]-N-(4-fluorophenyl)oxetane-3-carboxamide, Indoleamine 2,3-dioxygenase 1 | | Authors: | Lesburg, C.A. | | Deposit date: | 2021-08-09 | | Release date: | 2022-08-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Oxetane promise delivered: discovery of long acting IDO1 inhibitors suitable for Q3W oral or parenteral dosing
To Be Published
|
|
9GSX
 
 | | Campylobacter hook-filament junction-cap complex | | Descriptor: | Flagellar hook-associated protein 1, Flagellar hook-associated protein 2, Flagellin | | Authors: | Qin, K, Gonzalez-Rodriguez, N, Shmakova, E, Beeby, M, Bergeron, J.R.C. | | Deposit date: | 2024-09-16 | | Release date: | 2025-04-09 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | The structure of the complete extracellular bacterial flagellum reveals the mechanism of flagellin incorporation.
Nat Microbiol, 10, 2025
|
|
9GMZ
 
 | | CryoEM structure of PmcTnsC-dsDNA-AMPPNP in complex with PmcTnsAB hook | | Descriptor: | AAA+ ATPase domain-containing protein, DNA, Integrase, ... | | Authors: | Finocchio, G, Chanez, C, Querques, I, Speichert, K.J, Jinek, M. | | Deposit date: | 2024-08-30 | | Release date: | 2025-04-02 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of TnsC oligomerization and transposase recruitment in type I-B CRISPR-associated transposons.
Nucleic Acids Res., 53, 2025
|
|
7RWO
 
 | | Crystal Structure of BPTF bromodomain in complex with 4-chloro-2-methyl-5-[(1,2,3,4-tetrahydroisoquinolin-7-yl)amino]pyridazin-3(2H)-one | | Descriptor: | 1,2-ETHANEDIOL, 4-chloro-2-methyl-5-[(1,2,3,4-tetrahydroisoquinolin-7-yl)amino]pyridazin-3(2H)-one, Nucleosome-remodeling factor subunit BPTF | | Authors: | Zahid, H, Buchholz, C, Johnson, J.A, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2021-08-20 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition.
J.Med.Chem., 64, 2021
|
|
1MJA
 
 | | Crystal structure of yeast Esa1 histone acetyltransferase domain complexed with acetyl coenzyme A | | Descriptor: | COENZYME A, Esa1 protein | | Authors: | Yan, Y, Harper, S, Speicher, D, Marmorstein, R. | | Deposit date: | 2002-08-27 | | Release date: | 2002-10-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | The catalytic mechanism of the ESA1 histone acetyltransferase involves a self-acetylated intermediate.
Nat.Struct.Biol., 9, 2002
|
|
9FZB
 
 | | Human p53 DNA-binding domain bound to DARPin C10-H82R | | Descriptor: | 1,2-ETHANEDIOL, Cellular tumor antigen p53, DARPin C10-H82R, ... | | Authors: | Yuksel, B, Balourdas, D.I, Muenick, P, Knapp, S, Doetsch, V, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-07-05 | | Release date: | 2025-04-02 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | DARPin-induced reactivation of p53 in HPV-positive cells.
Nat.Struct.Mol.Biol., 32, 2025
|
|
1MJP
 
 | |
7RTZ
 
 | |
9G59
 
 | |
1MI2
 
 | | SOLUTION STRUCTURE OF MURINE MACROPHAGE INFLAMMATORY PROTEIN-2, NMR, 20 STRUCTURES | | Descriptor: | MACROPHAGE INFLAMMATORY PROTEIN-2 | | Authors: | Shao, W, Jerva, L.F, West, J, Lolis, E, Schweitzer, B.I. | | Deposit date: | 1997-10-24 | | Release date: | 1998-04-29 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure of murine macrophage inflammatory protein-2.
Biochemistry, 37, 1998
|
|
7RXO
 
 | | Crystal structure of CDK2 liganded with compound WN333 | | Descriptor: | 1,2-ETHANEDIOL, 2-{[2-(1H-indol-3-yl)ethyl]amino}-5-(methoxycarbonyl)benzoic acid, Cyclin-dependent kinase 2 | | Authors: | Sun, L, Schonbrunn, E. | | Deposit date: | 2021-08-23 | | Release date: | 2022-08-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Screening through Lead Optimization of High Affinity, Allosteric Cyclin-Dependent Kinase 2 (CDK2) Inhibitors as Male Contraceptives That Reduce Sperm Counts in Mice.
J.Med.Chem., 66, 2023
|
|
9G15
 
 | | Crystal structure of marine actinobacteria clade rhodopsin (MAR) in the O* state, pH 8.8 | | Descriptor: | Bacteriorhodopsin, EICOSANE, OLEIC ACID, ... | | Authors: | Bukhdruker, S, Kovalev, K, Astashkin, R, Gordeliy, V. | | Deposit date: | 2024-07-09 | | Release date: | 2025-04-02 | | Last modified: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Proteorhodopsin insights into the molecular mechanism of vectorial proton transport.
Sci Adv, 11, 2025
|
|
1LW7
 
 | | NADR PROTEIN FROM HAEMOPHILUS INFLUENZAE | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, TRANSCRIPTIONAL REGULATOR NADR | | Authors: | Singh, S.K, Kurnasov, O.V, Chen, B, Robinson, H, Grishin, N.V, Osterman, A.L, Zhang, H. | | Deposit date: | 2002-05-30 | | Release date: | 2002-08-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of Haemophilus influenzae NadR protein. A bifunctional enzyme endowed with NMN adenyltransferase and ribosylnicotinimide kinase activities.
J.Biol.Chem., 277, 2002
|
|
1MAY
 
 | | BETA-TRYPSIN PHOSPHONATE INHIBITED | | Descriptor: | BETA-TRYPSIN, CALCIUM ION, [N-(BENZYLOXYCARBONYL)AMINO](4-AMIDINOPHENYL)METHANE-PHOSPHONATE | | Authors: | Bertrand, J, Oleksyszyn, J, Kam, C, Boduszek, B, Presnell, S, Plaskon, R, Suddath, F, Powers, J, Williams, L. | | Deposit date: | 1996-02-06 | | Release date: | 1996-10-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Inhibition of trypsin and thrombin by amino(4-amidinophenyl)methanephosphonate diphenyl ester derivatives: X-ray structures and molecular models.
Biochemistry, 35, 1996
|
|
9G16
 
 | | Crystal structure of marine actinobacteria clade rhodopsin (MAR) in the O state obtained by cryotrapping | | Descriptor: | Bacteriorhodopsin, EICOSANE, OLEIC ACID, ... | | Authors: | Bukhdruker, S, Kovalev, K, Astashkin, R, Gordeliy, V. | | Deposit date: | 2024-07-09 | | Release date: | 2025-04-02 | | Last modified: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Proteorhodopsin insights into the molecular mechanism of vectorial proton transport.
Sci Adv, 11, 2025
|
|
