6CT9
 
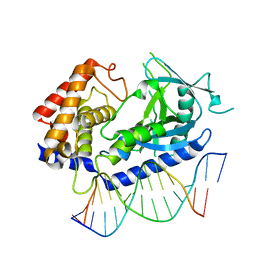 | | Structure of the human cGAS-DNA complex | | Descriptor: | Cyclic GMP-AMP synthase, DNA (5'-D(*AP*AP*AP*TP*TP*GP*CP*CP*GP*AP*AP*GP*AP*CP*GP*A)-3'), DNA (5'-D(P*CP*GP*TP*CP*TP*TP*CP*GP*GP*CP*AP*AP*T)-3'), ... | | Authors: | Zhou, W, Whiteley, A.T, de Oliveira Mann, C.C, Morehouse, B.R, Mekalanos, J.J, Kranzusch, P.J. | | Deposit date: | 2018-03-22 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structure of the Human cGAS-DNA Complex Reveals Enhanced Control of Immune Surveillance.
Cell, 174, 2018
|
|
3WKR
 
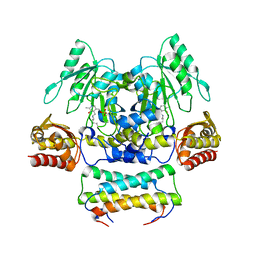 | | Crystal structure of the SepCysS-SepCysE complex from Methanocaldococcus jannaschii | | Descriptor: | O-phospho-L-seryl-tRNA:Cys-tRNA synthase, Uncharacterized protein MJ1481 | | Authors: | Nakazawa, Y, Asano, N, Nakamura, A, Yao, M. | | Deposit date: | 2013-10-30 | | Release date: | 2014-07-16 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Ancient translation factor is essential for tRNA-dependent cysteine biosynthesis in methanogenic archaea.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4GDM
 
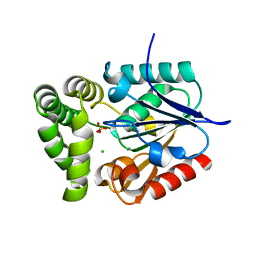 | | Crystal Structure of E.coli MenH | | Descriptor: | 2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylate synthase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Johnston, J.M, Baker, E.N, Guo, Z, Jiang, M. | | Deposit date: | 2012-07-31 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal Structures of E. coli Native MenH and Two Active Site Mutants.
Plos One, 8, 2013
|
|
1O23
 
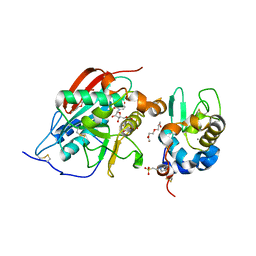 | | CRYSTAL STRUCTURE OF LACTOSE SYNTHASE IN THE PRESENCE OF UDP-GLUCOSE | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ALPHA-LACTALBUMIN, BETA-1,4-GALACTOSYLTRANSFERASE, ... | | Authors: | Ramakrishnan, B, Shah, P.S, Qasba, P.K. | | Deposit date: | 2003-01-29 | | Release date: | 2003-02-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Alpha-Lactalbumin (La) Stimulates Milk Beta-1,4-Galactosyltransferase I (Beta 4Gal-T1) to Transfer Glucose from Udp-Glucose to N-Acetylglucosamine. Crystal Structure of Beta 4Gal-T1 X La Complex with Udp-Glc.
J.Biol.Chem., 276, 2001
|
|
4GEG
 
 | | Crystal Structure of E.coli MenH Y85F Mutant | | Descriptor: | 1,2-ETHANEDIOL, 2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylate synthase, CHLORIDE ION, ... | | Authors: | Johnston, J.M, Baker, E.N, Guo, Z, Jiang, M. | | Deposit date: | 2012-08-01 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Crystal Structures of E. coli Native MenH and Two Active Site Mutants.
Plos One, 8, 2013
|
|
6MJW
 
 | | human cGAS catalytic domain bound with the inhibitor G150 | | Descriptor: | 1-[9-(6-aminopyridin-3-yl)-6,7-dichloro-1,3,4,5-tetrahydro-2H-pyrido[4,3-b]indol-2-yl]-2-hydroxyethan-1-one, Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Lama, L, Adura, C, Xie, W, Tomita, D, Kamei, T, Kuryavyi, V, Gogakos, T, Steinberg, J.I, Miller, M, Ramos-Espiritu, L, Asano, Y, Hashizume, S, Aida, J, Imaeda, T, Okamoto, R, Jennings, A.J, Michinom, M, Kuroita, T, Stamford, A, Gao, P, Meinke, P, Glickman, J.F, Patel, D.J, Tuschl, T. | | Deposit date: | 2018-09-23 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.405 Å) | | Cite: | Development of human cGAS-specific small-molecule inhibitors for repression of dsDNA-triggered interferon expression.
Nat Commun, 10, 2019
|
|
4GEC
 
 | | Crystal Structure of E.coli MenH R124A Mutant | | Descriptor: | 1,2-ETHANEDIOL, 2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylate synthase, CHLORIDE ION, ... | | Authors: | Johnston, J.M, Baker, E.N, Guo, Z, Jiang, M. | | Deposit date: | 2012-08-01 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structures of E. coli Native MenH and Two Active Site Mutants.
Plos One, 8, 2013
|
|
6MJU
 
 | | human cGAS catalytic domain bound with the inhibitor G108 | | Descriptor: | 1-[6,7-dichloro-9-(1H-pyrazol-4-yl)-1,3,4,5-tetrahydro-2H-pyrido[4,3-b]indol-2-yl]-2-hydroxyethan-1-one, Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Lama, L, Adura, C, Xie, W, Tomita, D, Kamei, T, Kuryavyi, V, Gogakos, T, Steinberg, J.I, Miller, M, Ramos-Espiritu, L, Asano, Y, Hashizume, S, Aida, J, Imaeda, T, Okamoto, R, Jennings, A.J, Michinom, M, Kuroita, T, Stamford, A, Gao, P, Meinke, P, Glickman, J.F, Patel, D.J, Tuschl, T. | | Deposit date: | 2018-09-22 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Development of human cGAS-specific small-molecule inhibitors for repression of dsDNA-triggered interferon expression.
Nat Commun, 10, 2019
|
|
6NAO
 
 | | Discovery of a high affinity inhibitor of cGAS | | Descriptor: | (1R,2S)-2-[(7-hydroxy-5-phenylpyrazolo[1,5-a]pyrimidine-3-carbonyl)amino]cyclohexane-1-carboxylic acid, CYCLIC GMP-AMP SYNTHASE, ZINC ION | | Authors: | Hall, J. | | Deposit date: | 2018-12-06 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.23 Å) | | Cite: | Discovery of PF-06928215 as a high affinity inhibitor of cGAS enabled by a novel fluorescence polarization assay.
PLoS ONE, 12, 2017
|
|
5V8O
 
 | | Discovery of a high affinity inhibitor of cGAS | | Descriptor: | 5-phenyltetrazolo[1,5-a]pyrimidin-7-ol, Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Hall, J. | | Deposit date: | 2017-03-22 | | Release date: | 2017-09-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Discovery of PF-06928215 as a high affinity inhibitor of cGAS enabled by a novel fluorescence polarization assay.
PLoS ONE, 12, 2017
|
|
1Q9J
 
 | | Structure of polyketide synthase associated protein 5 from Mycobacterium tuberculosis | | Descriptor: | Polyketide synthase associated protein 5 | | Authors: | Buglino, J, Onwueme, K.C, Quadri, L.E, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-08-25 | | Release date: | 2004-05-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal Structure of PapA5, a Phthiocerol Dimycocerosyl Transferase from Mycobacterium tuberculosis
J.Biol.Chem., 279, 2004
|
|
3FLM
 
 | | Crystal structure of menD from E.coli | | Descriptor: | 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylate synthase | | Authors: | Priyadarshi, A, Hwang, K.Y. | | Deposit date: | 2008-12-19 | | Release date: | 2009-03-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights of the MenD from Escherichia coli reveal ThDP affinity.
Biochem.Biophys.Res.Commun., 380, 2009
|
|
6B9S
 
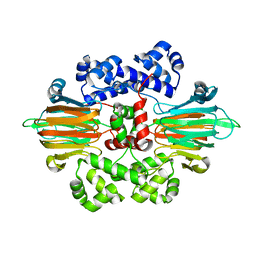 | |
6LKC
 
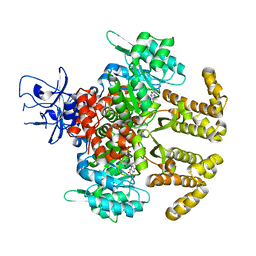 | | Crystal structure of PfaD from Shewanella piezotolerans in complex with FMN | | Descriptor: | CALCIUM ION, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Zhang, M.L, Li, Q, Meng, S.S, Guo, L.J, He, L, Huang, J.Z, Li, L, Zhang, H.D. | | Deposit date: | 2019-12-19 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Structural Insights into the Trans -Acting Enoyl Reductase in the Biosynthesis of Long-Chain Polyunsaturated Fatty Acids in Shewanella piezotolerans .
J.Agric.Food Chem., 69, 2021
|
|
6GMZ
 
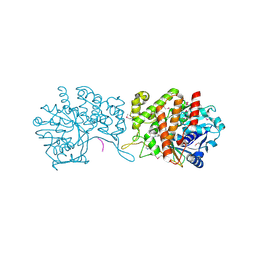 | |
1PDR
 
 | |
2QGI
 
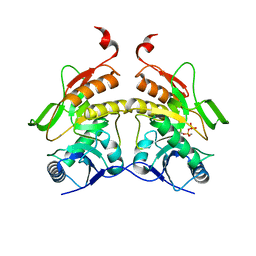 | |
7EQI
 
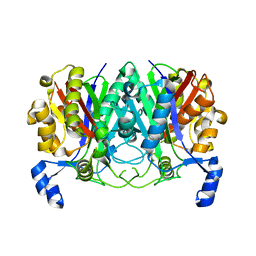 | | ChlB3 [Aceyltransferase] | | Descriptor: | ChlB3 | | Authors: | Saeed, A.U, Zheng, J. | | Deposit date: | 2021-05-02 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Insight of KSIII ( beta-Ketoacyl-ACP Synthase)-like Acyltransferase ChlB3 in the Biosynthesis of Chlorothricin.
Molecules, 27, 2022
|
|
3ROW
 
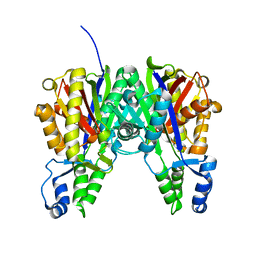 | | Crystal Structure of Xanthomonas campestri OleA | | Descriptor: | 3-oxoacyl-[ACP] synthase III | | Authors: | Goblirsch, B.R, Wilmot, C.M. | | Deposit date: | 2011-04-26 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8487 Å) | | Cite: | Crystal Structures of Xanthomonas campestris OleA Reveal Features That Promote Head-to-Head Condensation of Two Long-Chain Fatty Acids.
Biochemistry, 51, 2012
|
|
3S1Z
 
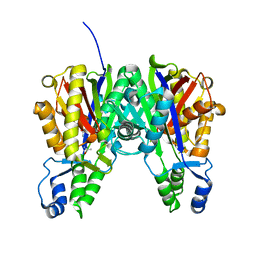 | |
3S20
 
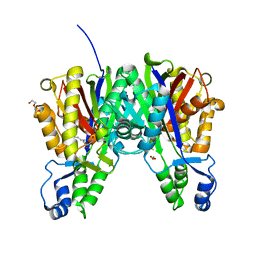 | | Crystal structure of cerulenin bound Xanthomonas campestri OleA (soak) | | Descriptor: | (2S, 3R)-3-HYDROXY-4-OXO-7,10-TRANS,TRANS-DODECADIENAMIDE, 3-oxoacyl-[ACP] synthase III, ... | | Authors: | Goblirsch, B.R, Wilmot, C.M. | | Deposit date: | 2011-05-16 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8796 Å) | | Cite: | Crystal Structures of Xanthomonas campestris OleA Reveal Features That Promote Head-to-Head Condensation of Two Long-Chain Fatty Acids.
Biochemistry, 51, 2012
|
|
1PJS
 
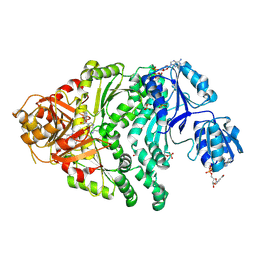 | | The co-crystal structure of CysG, the multifunctional methyltransferase/dehydrogenase/ferrochelatase for siroheme synthesis, in complex with it NAD cofactor | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PHOSPHATE ION, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Stroupe, M.E, Leech, H.K, Daniels, D.S, Warren, M.J, Getzoff, E.D. | | Deposit date: | 2003-06-03 | | Release date: | 2003-12-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | CysG structure reveals tetrapyrrole-binding features and novel regulation of siroheme biosynthesis.
Nat.Struct.Biol., 10, 2003
|
|
1PJT
 
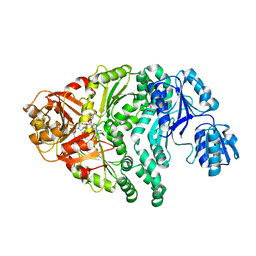 | | The structure of the Ser128Ala point-mutant variant of CysG, the multifunctional methyltransferase/dehydrogenase/ferrochelatase for siroheme synthesis | | Descriptor: | PHOSPHATE ION, S-ADENOSYL-L-HOMOCYSTEINE, Siroheme synthase | | Authors: | Stroupe, M.E, Leech, H.K, Daniels, D.S, Warren, M.J, Getzoff, E.D. | | Deposit date: | 2003-06-03 | | Release date: | 2003-12-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | CysG structure reveals tetrapyrrole-binding features and novel regulation of siroheme biosynthesis.
Nat.Struct.Biol., 10, 2003
|
|
7W46
 
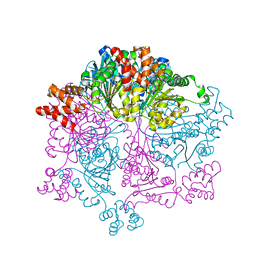 | | Crystal structure of Bacillus subtilis YjoB with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Uncharacterized ATPase YjoB | | Authors: | Dahal, P, Kwon, E, Kim, D.Y. | | Deposit date: | 2021-11-26 | | Release date: | 2022-10-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure and biochemical analysis suggest that YjoB ATPase is a putative substrate-specific molecular chaperone.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7W42
 
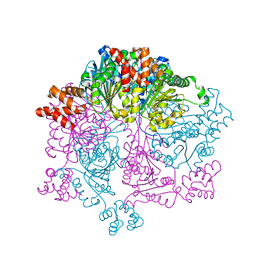 | | Crystal structure of Bacillus subtilis YjoB | | Descriptor: | Uncharacterized ATPase YjoB | | Authors: | Dahal, P, Kwon, E, Kim, D.Y. | | Deposit date: | 2021-11-26 | | Release date: | 2022-10-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.619 Å) | | Cite: | Crystal structure and biochemical analysis suggest that YjoB ATPase is a putative substrate-specific molecular chaperone.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
