5OR3
 
 | | Crystal structure of Aspergillus oryzae catechol oxidase in met/deoxy-form | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hakulinen, N, Penttinen, L, Rutanen, C, Rouvinen, J. | | Deposit date: | 2017-08-15 | | Release date: | 2018-05-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.795 Å) | | Cite: | A new crystal form of Aspergillus oryzae catechol oxidase and evaluation of copper site structures in coupled binuclear copper enzymes.
PLoS ONE, 13, 2018
|
|
3SMH
 
 | |
5OAR
 
 | | Crystal structure of native beta-N-acetylhexosaminidase isolated from Aspergillus oryzae | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, ... | | Authors: | Skerlova, J, Rezacova, P, Brynda, J, Pachl, P, Otwinowski, Z, Vanek, O. | | Deposit date: | 2017-06-23 | | Release date: | 2017-12-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of native beta-N-acetylhexosaminidase isolated from Aspergillus oryzae sheds light onto its substrate specificity, high stability, and regulation by propeptide.
FEBS J., 285, 2018
|
|
3UTE
 
 | | Crystal structure of Aspergillus fumigatus UDP galactopyranose mutase sulfate complex | | Descriptor: | ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Dhatwalia, R, Singh, H, Tanner, J.J. | | Deposit date: | 2011-11-25 | | Release date: | 2012-02-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structures and Small-angle X-ray Scattering Analysis of UDP-galactopyranose Mutase from the Pathogenic Fungus Aspergillus fumigatus.
J.Biol.Chem., 287, 2012
|
|
3UTF
 
 | |
3UTG
 
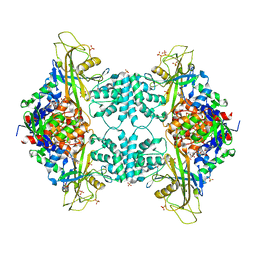 | | Crystal structure of Aspergillus fumigatus UDP galactopyranose mutase complexed with UDP in reduced state | | Descriptor: | DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, SULFATE ION, UDP-galactopyranose mutase, ... | | Authors: | Dhatwalia, R, Singh, H, Tanner, J.J. | | Deposit date: | 2011-11-25 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structures and Small-angle X-ray Scattering Analysis of UDP-galactopyranose Mutase from the Pathogenic Fungus Aspergillus fumigatus.
J.Biol.Chem., 287, 2012
|
|
3UTH
 
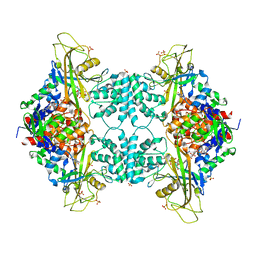 | | Crystal structure of Aspergillus fumigatus UDP galactopyranose mutase complexed with substrate UDP-Galp in reduced state | | Descriptor: | DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, GALACTOSE-URIDINE-5'-DIPHOSPHATE, SULFATE ION, ... | | Authors: | Dhatwalia, R, Singh, H, Tanner, J.J. | | Deposit date: | 2011-11-25 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structures and Small-angle X-ray Scattering Analysis of UDP-galactopyranose Mutase from the Pathogenic Fungus Aspergillus fumigatus.
J.Biol.Chem., 287, 2012
|
|
3PRG
 
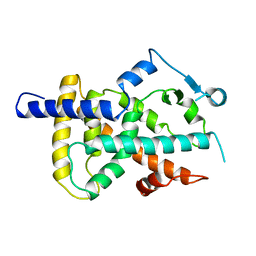 | | LIGAND BINDING DOMAIN OF HUMAN PEROXISOME PROLIFERATOR ACTIVATED RECEPTOR | | Descriptor: | PEROXISOME PROLIFERATOR ACTIVATED RECEPTOR GAMMA | | Authors: | Uppenberg, J, Svensson, C, Jaki, M, Bertilsson, G, Jendeberg, L, Berkenstam, A. | | Deposit date: | 1998-08-24 | | Release date: | 1999-08-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the ligand binding domain of the human nuclear receptor PPARgamma.
J.Biol.Chem., 273, 1998
|
|
4A23
 
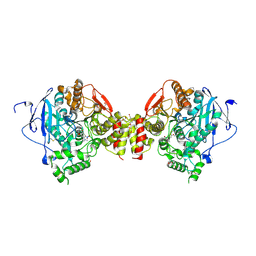 | | Mus musculus Acetylcholinesterase in complex with racemic C5685 | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 4-(DIMETHYLAMINO)-N-{[(2R)-1-ETHYLPYRROLIDIN-2-YL]METHYL}-2-METHOXY-5-NITROBENZAMIDE, 4-(DIMETHYLAMINO)-N-{[(2S)-1-ETHYLPYRROLIDIN-2-YL]METHYL}-2-METHOXY-5-NITROBENZAMIDE, ... | | Authors: | Berg, L, Andersson, C.D, Artrursson, E, Hornberg, A, Tunemalm, A.K, Linusson, A, Ekstrom, F. | | Deposit date: | 2011-09-22 | | Release date: | 2011-12-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Targeting Acetylcholinesterase: Identification of Chemical Leads by High Throughput Screening, Structure Determination and Molecular Modeling.
Plos One, 6, 2011
|
|
4QNW
 
 | |
3RJA
 
 | | Crystal structure of carbohydrate oxidase from Microdochium nivale in complex with substrate analogue | | Descriptor: | (2R,3R,4R,5R)-4,5-dihydroxy-2-(hydroxymethyl)-6-oxopiperidin-3-yl beta-D-glucopyranoside, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Duskova, J, Skalova, T, Kolenko, P, Stepankova, A, Koval, T, Hasek, J, Ostergaard, L.H, Fuglsang, C.C, Dohnalek, J. | | Deposit date: | 2011-04-15 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure and kinetic studies of carbohydrate oxidase from Microdochium nivale
To be Published
|
|
3RJ8
 
 | | Crystal structure of carbohydrate oxidase from Microdochium nivale | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Carbohydrate oxidase, ... | | Authors: | Duskova, J, Skalova, T, Stepankova, A, Koval, T, Hasek, J, Ostergaard, L.H, Fuglsang, C.C, Kolenko, P, Dohnalek, J. | | Deposit date: | 2011-04-15 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure and kinetic studies of carbohydrate oxidase from Microdochium nivale
To be Published
|
|
3RWK
 
 | | First crystal structure of an endo-inulinase, from Aspergillus ficuum: structural analysis and comparison with other GH32 enzymes. | | Descriptor: | ACETATE ION, Inulinase, SODIUM ION, ... | | Authors: | Michaux, C, Pouyez, J, Roussel, G, Mayard, A, Vandamme, A.M, Housen, I, Wouters, J. | | Deposit date: | 2011-05-09 | | Release date: | 2012-07-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | First crystal structure of an endo-inulinase, INU2, from Aspergillus ficuum: Discovery of an extra-pocket in the catalytic domain responsible for its endo-activity.
Biochimie, 94, 2012
|
|
3SC7
 
 | | First crystal structure of an endo-inulinase, from Aspergillus ficuum: structural analysis and comparison with other GH32 enzymes. | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Inulinase, alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose, ... | | Authors: | Housen, I, Pouyez, J, Roussel, G, Mayard, A, Vandamme, A.M, Wouters, J, Michaux, C. | | Deposit date: | 2011-06-07 | | Release date: | 2012-06-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | First crystal structure of an endo-inulinase, INU2, from Aspergillus ficuum: Discovery of an extra-pocket in the catalytic domain responsible for its endo-activity.
Biochimie, 94, 2012
|
|
4EL9
 
 | | Structure of N-terminal kinase domain of RSK2 with afzelin | | Descriptor: | 5,7-dihydroxy-2-(4-hydroxyphenyl)-4-oxo-4H-chromen-3-yl 6-deoxy-alpha-L-mannopyranoside, Ribosomal protein S6 kinase alpha-3 | | Authors: | Utepbergenov, D, Derewenda, U, Derewenda, Z.S. | | Deposit date: | 2012-04-10 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Insights into the Inhibition of the p90 Ribosomal S6 Kinase (RSK) by the Flavonol Glycoside SL0101 from the 1.5 A Crystal Structure of the N-Terminal Domain of RSK2 with Bound Inhibitor.
Biochemistry, 51, 2012
|
|
3UKP
 
 | |
3UKH
 
 | |
7JJO
 
 | | Structural Basis of the Activation of Heterotrimeric Gs-protein by Isoproterenol-bound Beta1-Adrenergic Receptor | | Descriptor: | Beta1-Adrenergic Receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Su, M, Zhu, L, Zhang, Y, Paknejad, N, Dey, R, Huang, J, Lee, M.Y, Williams, D, Jordan, K.D, Eng, E.T, Ernst, O.P, Meyerson, J.R, Hite, R.K, Walz, T, Liu, W, Huang, X.Y. | | Deposit date: | 2020-07-27 | | Release date: | 2020-09-02 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural Basis of the Activation of Heterotrimeric Gs-Protein by Isoproterenol-Bound beta 1 -Adrenergic Receptor.
Mol.Cell, 80, 2020
|
|
4QBJ
 
 | | Crystal structure of N-myristoyl transferase from Aspergillus fumigatus complexed with a synthetic inhibitor | | Descriptor: | 3-[[3-methyl-2-[[2,3,4-tris(fluoranyl)phenoxy]methyl]-1-benzofuran-4-yl]oxy]-N-(pyridin-3-ylmethyl)propan-1-amine, Glycylpeptide N-tetradecanoyltransferase, S-(2-OXO)PENTADECYLCOA, ... | | Authors: | Suzuki, M, Shimada, T. | | Deposit date: | 2014-05-08 | | Release date: | 2015-04-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of N-myristoyltransferase from Aspergillus fumigatus
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3UBD
 
 | | Structure of N-terminal domain of RSK2 kinase in complex with flavonoid glycoside SL0101 | | Descriptor: | 5,7-dihydroxy-2-(4-hydroxyphenyl)-4-oxo-4H-chromen-3-yl 3,4-di-O-acetyl-6-deoxy-alpha-L-mannopyranoside, Ribosomal protein S6 kinase alpha-3 | | Authors: | Utepbergenov, D, Derewenda, U, Derewenda, Z.S. | | Deposit date: | 2011-10-24 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Insights into the Inhibition of the p90 Ribosomal S6 Kinase (RSK) by the Flavonol Glycoside SL0101 from the 1.5 A Crystal Structure of the N-Terminal Domain of RSK2 with Bound Inhibitor.
Biochemistry, 51, 2012
|
|
3UKF
 
 | |
4HW4
 
 | |
4OY8
 
 | | Structure of ScLPMO10B in complex with zinc. | | Descriptor: | ACETATE ION, Putative secreted cellulose-binding protein, ZINC ION | | Authors: | Forsberg, Z, Mackenzie, A.K, Sorlie, M, Rohr, A.K, Helland, R, Arvai, A.S, Vaaje-Kolstad, G, Eijsink, V.G.H. | | Deposit date: | 2014-02-11 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and functional characterization of a conserved pair of bacterial cellulose-oxidizing lytic polysaccharide monooxygenases.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3NSZ
 
 | | Human CK2 catalytic domain in complex with AMPPN | | Descriptor: | Casein kinase II subunit alpha, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Ferguson, A.D. | | Deposit date: | 2010-07-02 | | Release date: | 2010-12-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis of CX-4945 binding to human protein kinase CK2.
Febs Lett., 585, 2011
|
|
7KOT
 
 | |
