5EVM
 
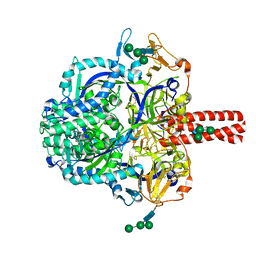 | | Crystal Structure of Nipah Virus Fusion Glycoprotein in the Prefusion State | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fusion glycoprotein F0, MALONATE ION, ... | | Authors: | Xu, K, Nikolov, D.B. | | Deposit date: | 2015-11-20 | | Release date: | 2015-12-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.367 Å) | | Cite: | Crystal Structure of the Pre-fusion Nipah Virus Fusion Glycoprotein Reveals a Novel Hexamer-of-Trimers Assembly.
Plos Pathog., 11, 2015
|
|
1Z4Z
 
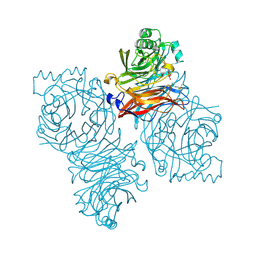 | | Parainfluenza Virus 5 (SV5) Hemagglutinin-Neuraminidase (HN) with ligand DANA(soaked with sialic acid, pH7.0)) | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yuan, P, Thompson, T.B, Wurzburg, B.A, Paterson, R.G, Lamb, R.A, Jardetzky, T.S. | | Deposit date: | 2005-03-16 | | Release date: | 2005-05-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural studies of the parainfluenza virus 5 hemagglutinin-neuraminidase tetramer in complex with its receptor, sialyllactose.
Structure, 13, 2005
|
|
5EXN
 
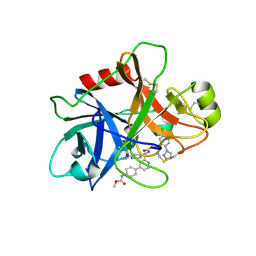 | | FACTOR XIA (C500S [C122S]) IN COMPLEX WITH THE INHIBITOR methyl ~{N}-[4-[2-[(1~{S})-1-[[(~{E})-3-[5-chloranyl-2-(1,2,3,4-tetrazol-1-yl)phenyl]prop-2-enoyl]amino]-2-phenyl-ethyl]pyridin-4-yl]phenyl]carbamate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Coagulation factor XIa light chain, methyl ~{N}-[4-[2-[(1~{S})-1-[[(~{E})-3-[5-chloranyl-2-(1,2,3,4-tetrazol-1-yl)phenyl]prop-2-enoyl]amino]-2-phenyl-ethyl]pyridin-4-yl]phenyl]carbamate | | Authors: | Sheriff, S. | | Deposit date: | 2015-11-23 | | Release date: | 2016-04-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Orally bioavailable pyridine and pyrimidine-based Factor XIa inhibitors: Discovery of the methyl N-phenyl carbamate P2 prime group
Bioorg.Med.Chem., 24, 2016
|
|
8IA7
 
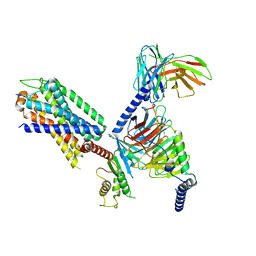 | | Structural insights into human brain gut peptide cholecystokinin receptors | | Descriptor: | CCK-8, Gastrin/cholecystokinin type B receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Ding, Y, Zhang, H, Liao, Y, Chen, L, Ji, S. | | Deposit date: | 2023-02-08 | | Release date: | 2023-12-06 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into human brain-gut peptide cholecystokinin receptors.
Cell Discov, 8, 2022
|
|
8PEP
 
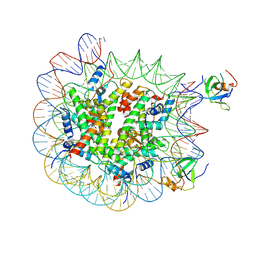 | | H3K36me2 nucleosome-LEDGF/p75 PWWP domain complex - pose 2 | | Descriptor: | Histone H2A, Histone H2B 1.1, Histone H3, ... | | Authors: | Koutna, E, Kouba, T, Veverka, V. | | Deposit date: | 2023-06-14 | | Release date: | 2023-08-16 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Multivalency of nucleosome recognition by LEDGF.
Nucleic Acids Res., 51, 2023
|
|
8PC5
 
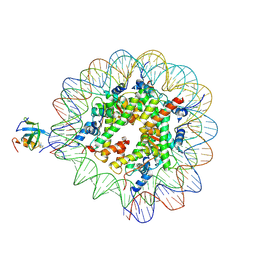 | | H3K36me3 nucleosome-LEDGF/p75 PWWP domain complex | | Descriptor: | Histone H2A, Histone H2B 1.1, Histone H3, ... | | Authors: | Koutna, E, Kouba, T, Veverka, V. | | Deposit date: | 2023-06-09 | | Release date: | 2023-08-16 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Multivalency of nucleosome recognition by LEDGF.
Nucleic Acids Res., 51, 2023
|
|
8PEO
 
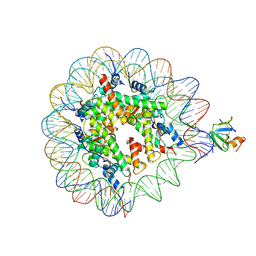 | | H3K36me2 nucleosome-LEDGF/p75 PWWP domain complex | | Descriptor: | Histone H2A, Histone H2B 1.1, Histone H3, ... | | Authors: | Koutna, E, Kouba, T, Veverka, V. | | Deposit date: | 2023-06-14 | | Release date: | 2023-08-16 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Multivalency of nucleosome recognition by LEDGF.
Nucleic Acids Res., 51, 2023
|
|
8PC6
 
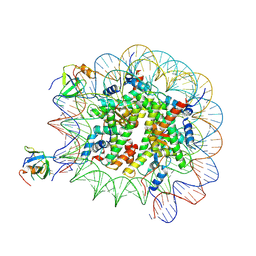 | | H3K36me3 nucleosome-LEDGF/p75 PWWP domain complex - pose 2 | | Descriptor: | Histone H2A, Histone H2B 1.1, Histone H3, ... | | Authors: | Koutna, E, Kouba, T, Veverka, V. | | Deposit date: | 2023-06-09 | | Release date: | 2023-08-16 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Multivalency of nucleosome recognition by LEDGF.
Nucleic Acids Res., 51, 2023
|
|
2PES
 
 | | Urate Oxidase in complex with tris-dipicolinate Lutetium | | Descriptor: | 8-AZAXANTHINE, LUTETIUM (III) ION, PYRIDINE-2,6-DICARBOXYLIC ACID, ... | | Authors: | Pompidor, G, Vicat, J, Kahn, R. | | Deposit date: | 2007-04-03 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A dipicolinate lanthanide complex for solving protein structures using anomalous diffraction.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
5F4E
 
 | | Crystal structure of the human sperm Izumo1 and egg Juno complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLYCEROL, ... | | Authors: | Aydin, H, Sultana, A, Lee, J.E. | | Deposit date: | 2015-12-03 | | Release date: | 2016-06-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular architecture of the human sperm IZUMO1 and egg JUNO fertilization complex.
Nature, 534, 2016
|
|
5F4V
 
 | |
8PG0
 
 | | Human OATP1B3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BICARBONATE ION, CHOLESTEROL, ... | | Authors: | Ciuta, A.-D, Nosol, K, Kowal, J, Mukherjee, S, Ramirez, A.S, Stieger, B, Kossiakoff, A.A, Locher, K.P. | | Deposit date: | 2023-06-17 | | Release date: | 2023-09-27 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structure of human drug transporters OATP1B1 and OATP1B3.
Nat Commun, 14, 2023
|
|
5EWS
 
 | | Sugar binding protein - human galectin-2 | | Descriptor: | Galectin-2, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Su, J.Y, Si, Y.L. | | Deposit date: | 2015-11-21 | | Release date: | 2016-09-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human galectin-2 interacts with carbohydrates and peptides non-classically: new insight from X-ray crystallography and hemagglutination.
Acta Biochim. Biophys. Sin. (Shanghai), 48, 2016
|
|
1Z0M
 
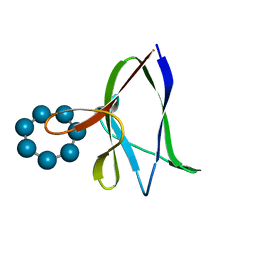 | | the glycogen-binding domain of the AMP-activated protein kinase beta1 subunit | | Descriptor: | 5'-AMP-activated protein kinase, beta-1 subunit, Cycloheptakis-(1-4)-(alpha-D-glucopyranose) | | Authors: | Polekhina, G, Gupta, A, van Denderen, B.J, Feil, S.C, Kemp, B.E, Stapleton, D, Parker, M.W. | | Deposit date: | 2005-03-02 | | Release date: | 2005-10-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural Basis for Glycogen Recognition by AMP-Activated Protein Kinase.
Structure, 13, 2005
|
|
5F09
 
 | | Structure of inactive GCPII mutant in complex with beta-citryl glutamate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Tykvart, J, Navratil, M, Pachl, P, Konvalinka, J. | | Deposit date: | 2015-11-27 | | Release date: | 2016-06-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Comparison of human glutamate carboxypeptidases II and III reveals their divergent substrate specificities.
Febs J., 283, 2016
|
|
1Z4W
 
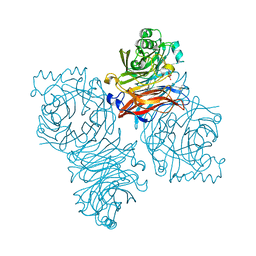 | | Parainfluenza Virus 5 (SV5) Hemagglutinin-Neuraminidase (HN) with ligand DANA (soaked with DANA, pH8.0) | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yuan, P, Thompson, T.B, Wurzburg, B.A, Paterson, R.G, Lamb, R.A, Jardetzky, T.S. | | Deposit date: | 2005-03-16 | | Release date: | 2005-05-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural studies of the parainfluenza virus 5 hemagglutinin-neuraminidase tetramer in complex with its receptor, sialyllactose.
Structure, 13, 2005
|
|
8PNW
 
 | | Crystal structure of D-amino acid aminotransferase from Blastococcus saxobsidens in holo form with PLP | | Descriptor: | Branched-chain amino acid aminotransferase/4-amino-4-deoxychorismate lyase, CHLORIDE ION, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Matyuta, I.O, Boyko, K.M, Nikolaeva, A.Y, Shilova, S.A, Popov, V.O. | | Deposit date: | 2023-07-03 | | Release date: | 2023-10-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Expanded Substrate Specificity in D-Amino Acid Transaminases: A Case Study of Transaminase from Blastococcus saxobsidens.
Int J Mol Sci, 24, 2023
|
|
5EW1
 
 | | Human thrombin sandwiched between two DNA aptamers: HD22 and HD1-deltaT3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, HD1-deltaT3, ... | | Authors: | Pica, A, Russo Krauss, I, Parente, V, Sica, F. | | Deposit date: | 2015-11-20 | | Release date: | 2016-11-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Through-bond effects in the ternary complexes of thrombin sandwiched by two DNA aptamers.
Nucleic Acids Res., 45, 2017
|
|
5EWL
 
 | |
8PXJ
 
 | | Structure of Whitewater Arroyo virus GP1 glycoprotein, solved at wavelength 2.75 A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CADMIUM ION, Glycoprotein G1, ... | | Authors: | El Omari, K, Duman, R, Mykhaylyk, V, Orr, C, Bowden, T.A, Wagner, A. | | Deposit date: | 2023-07-23 | | Release date: | 2023-10-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
8PNY
 
 | | Crystal structure of D-amino acid aminotransferase from Blastococcus saxobsidens complexed with phenylhydrazine and in its apo form | | Descriptor: | Branched-chain amino acid aminotransferase/4-amino-4-deoxychorismate lyase, [6-methyl-5-oxidanyl-4-[(2-phenylhydrazinyl)methyl]pyridin-3-yl]methyl dihydrogen phosphate | | Authors: | Matyuta, I.O, Boyko, K.M, Nikolaeva, A.Y, Shilova, S.A, Popov, V.O. | | Deposit date: | 2023-07-03 | | Release date: | 2023-10-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Expanded Substrate Specificity in D-Amino Acid Transaminases: A Case Study of Transaminase from Blastococcus saxobsidens.
Int J Mol Sci, 24, 2023
|
|
8PQ2
 
 | | XBB 1.0 RBD bound to P4J15 (Local) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, P4J15 Fragment Antigen-Binding Heavy Chain, P4J15 Fragment Antigen-Binding Light Chain, ... | | Authors: | Duhoo, Y, Lau, K. | | Deposit date: | 2023-07-10 | | Release date: | 2023-11-01 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Broadly potent anti-SARS-CoV-2 antibody shares 93% of epitope with ACE2 and provides full protection in monkeys.
J Infect, 87, 2023
|
|
8PI1
 
 | | Bicyclic INCYPRO Pseudomonas fluorescens esterase | | Descriptor: | Arylesterase, GLYCEROL, N-[2-[3,5-bis[2-(2-iodanylethanoylamino)ethanoyl]-1,3,5-triazinan-1-yl]-2-oxidanylidene-ethyl]-2-iodanyl-ethanamide | | Authors: | Kiehstaller, S, Pearce, N.M, Grossmann, T.N, Hennig, S. | | Deposit date: | 2023-06-20 | | Release date: | 2023-11-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Covalent bicyclization of protein complexes yields durable quaternary structures.
Chem, 10, 2024
|
|
8PSD
 
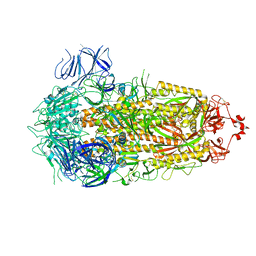 | | SARS-CoV-2 XBB 1.0 closed conformation. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Duhoo, Y, Lau, K. | | Deposit date: | 2023-07-13 | | Release date: | 2023-11-01 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Broadly potent anti-SARS-CoV-2 antibody shares 93% of epitope with ACE2 and provides full protection in monkeys.
J Infect, 87, 2023
|
|
5EW2
 
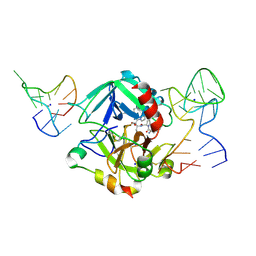 | | Human thrombin sandwiched between two DNA aptamers: HD22 and HD1-deltaT12 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, HD1-deltaT12, ... | | Authors: | Pica, A, Russo Krauss, I, Parente, V, Sica, F. | | Deposit date: | 2015-11-20 | | Release date: | 2016-11-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | Through-bond effects in the ternary complexes of thrombin sandwiched by two DNA aptamers.
Nucleic Acids Res., 45, 2017
|
|
