6ZBI
 
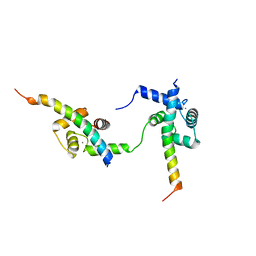 | | Ternary complex of Calmodulin bound to 2 molecules of NHE1 | | Descriptor: | CALCIUM ION, Calmodulin-1, Sodium/hydrogen exchanger 1 | | Authors: | Prestel, A, Kragelund, B.B, Pedersen, E.S, Pedersen, S.F, Sjoegaard-Frich, L.M. | | Deposit date: | 2020-06-08 | | Release date: | 2021-03-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Dynamic Na + /H + exchanger 1 (NHE1) - calmodulin complexes of varying stoichiometry and structure regulate Ca 2+ -dependent NHE1 activation.
Elife, 10, 2021
|
|
8OX2
 
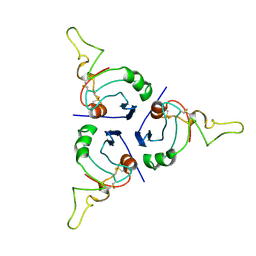 | | BRICHOS trimer | | Descriptor: | Pulmonary surfactant-associated protein C | | Authors: | Ghosh, D, Torres, F, Guentert, P, Riek, R. | | Deposit date: | 2023-04-30 | | Release date: | 2024-06-26 | | Last modified: | 2024-12-04 | | Method: | SOLUTION NMR | | Cite: | The inhibitory action of the chaperone BRICHOS against the alpha-Synuclein secondary nucleation pathway.
Nat Commun, 15, 2024
|
|
8OVI
 
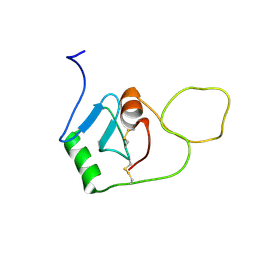 | | BRICHOS monomer | | Descriptor: | Pulmonary surfactant-associated protein C (Fragment) | | Authors: | Ghosh, D, Torres, F, Guentert, P, Riek, R. | | Deposit date: | 2023-04-26 | | Release date: | 2024-06-26 | | Last modified: | 2024-12-04 | | Method: | SOLUTION NMR | | Cite: | The inhibitory action of the chaperone BRICHOS against the alpha-Synuclein secondary nucleation pathway.
Nat Commun, 15, 2024
|
|
8PP5
 
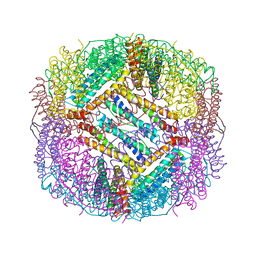 | |
8PP4
 
 | |
8PP3
 
 | |
6S7T
 
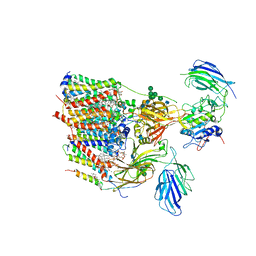 | | Cryo-EM structure of human oligosaccharyltransferase complex OST-B | | Descriptor: | (2Z,6Z,10Z,14Z,18Z,22Z,26Z)-3,7,11,15,19,23,27,31-octamethyldotriaconta-2,6,10,14,18,22,26,30-octaen-1-yl dihydrogen phosphate, (2~{S},3~{R},4~{R},5~{S},6~{S})-2-(hydroxymethyl)-6-[(1~{S},2~{R},3~{R},4~{R},5'~{S},6~{S},7~{R},8~{S},9~{R},12~{R},13~{R},15~{S},16~{S},18~{R})-5',7,9,13-tetramethyl-3,15-bis(oxidanyl)spiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icosane-6,2'-oxane]-16-yl]oxy-oxane-3,4,5-triol, (4R,7R)-4-hydroxy-N,N,N-trimethyl-4,9-dioxo-7-[(undecanoyloxy)methyl]-3,5,8-trioxa-4lambda~5~-phosphadocosan-1-aminium, ... | | Authors: | Ramirez, A.S, Kowal, J, Locher, K.P. | | Deposit date: | 2019-07-05 | | Release date: | 2019-12-18 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-electron microscopy structures of human oligosaccharyltransferase complexes OST-A and OST-B.
Science, 366, 2019
|
|
1XKD
 
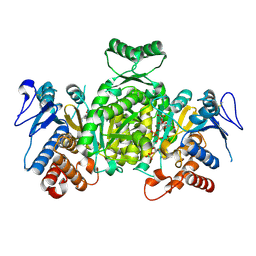 | | Ternary complex of Isocitrate dehydrogenase from the hyperthermophile Aeropyrum pernix | | Descriptor: | CALCIUM ION, ISOCITRIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Karlstrom, M, Stokke, R, Steen, I.H, Birkeland, N.-K, Ladenstein, R. | | Deposit date: | 2004-09-28 | | Release date: | 2005-10-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Isocitrate dehydrogenase from the hyperthermophile Aeropyrum pernix: X-ray structure analysis of a ternary enzyme-substrate complex and thermal stability.
J.Mol.Biol., 345, 2005
|
|
5OGL
 
 | | Structure of bacterial oligosaccharyltransferase PglB in complex with an acceptor peptide and an lipid-linked oligosaccharide analog | | Descriptor: | MANGANESE (II) ION, SODIUM ION, Substrate mimicking peptide, ... | | Authors: | Napiorkowska, M, Boilevin, J, Sovdat, T, Darbre, T, Reymond, J.-L, Aebi, M, Locher, K.P. | | Deposit date: | 2017-07-13 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular basis of lipid-linked oligosaccharide recognition and processing by bacterial oligosaccharyltransferase.
Nat. Struct. Mol. Biol., 24, 2017
|
|
1TNM
 
 | |
8PI1
 
 | | Bicyclic INCYPRO Pseudomonas fluorescens esterase | | Descriptor: | Arylesterase, GLYCEROL, N-[2-[3,5-bis[2-(2-iodanylethanoylamino)ethanoyl]-1,3,5-triazinan-1-yl]-2-oxidanylidene-ethyl]-2-iodanyl-ethanamide | | Authors: | Kiehstaller, S, Pearce, N.M, Grossmann, T.N, Hennig, S. | | Deposit date: | 2023-06-20 | | Release date: | 2023-11-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Covalent bicyclization of protein complexes yields durable quaternary structures.
Chem, 10, 2024
|
|
7Q2J
 
 | | Quaternary Complex of human WDR5 and pVHL:ElonginC:ElonginB bound to PROTAC Homer | | Descriptor: | Elongin-B, Elongin-C, N-[5-[4-[[5-[[(2S)-3,3-dimethyl-1-[(2S,4R)-2-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methylcarbamoyl]-4-oxidanyl-pyrrolidin-1-yl]-1-oxidanylidene-butan-2-yl]amino]-5-oxidanylidene-pentyl]carbamoyl]phenyl]-2-(4-methylpiperazin-1-yl)phenyl]-6-oxidanylidene-4-(trifluoromethyl)-1H-pyridine-3-carboxamide, ... | | Authors: | Kraemer, A, Doelle, A, Schwalm, M.P, Adhikari, B, Wolf, E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-10-25 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Tracking the PROTAC degradation pathway in living cells highlights the importance of ternary complex measurement for PROTAC optimization.
Cell Chem Biol, 30, 2023
|
|
2OH2
 
 | | Ternary Complex of Human DNA Polymerase | | Descriptor: | 5'-D(*GP*GP*G*GP*GP*AP*AP*GP*GP*AP*CP*CP*C)-3', 5'-D(*TP*T*CP*CP*AP*GP*GP*GP*TP*CP*CP*TP*TP*CP*CP*CP*CP*C)-3', DNA polymerase kappa, ... | | Authors: | Lone, S, Townson, S.A, Uljon, S.N, Prakash, S, Prakash, L, Aggarwal, A.K. | | Deposit date: | 2007-01-09 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Ternary complex of the catalytic core of human DNA polymerase Kappa with DNA and dTT
To be Published
|
|
5BPK
 
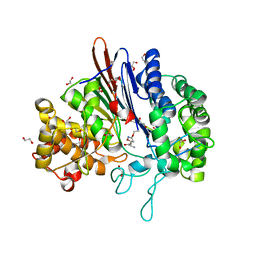 | | Varying binding modes of inhibitors and structural differences in the binding pockets of different gamma-glutamyltranspeptidases | | Descriptor: | (2S)-amino[(5S)-4,5-dihydro-1,2-oxazol-5-yl]acetic acid, 1,2-ETHANEDIOL, Gamma-glutamyltranspeptidase (Ggt) | | Authors: | Bolz, C, Bach, N.C, Meyer, H, Mueller, G, Dawidowski, M, Popowicz, G, Sieber, S.A, Skerra, A, Gerhard, M. | | Deposit date: | 2015-05-28 | | Release date: | 2016-05-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Varying binding modes of inhibitors and structural differences in the binding pockets of different gamma-glutamyltranspeptidases
To Be Published
|
|
1HBX
 
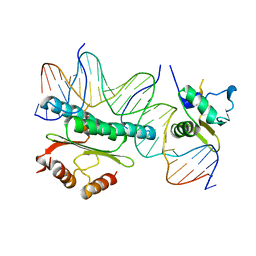 | | Ternary Complex of SAP-1 and SRF with specific SRE DNA | | Descriptor: | 5'-D(*CP*AP*CP*AP*CP*CP*GP*GP*AP*AP*GP*TP*CP* CP*TP*AP*AP*TP*TP*AP*GP*GP*CP*CP*AP*T)-3', 5'-D(*GP*AP*TP*GP*GP*CP*CP*TP*AP*AP*TP*TP*AP* GP*GP*AP*CP*TP*TP*CP*CP*GP*GP*TP*G)-3', ETS-DOMAIN PROTEIN ELK-4, ... | | Authors: | Hassler, M, Richmond, T.J. | | Deposit date: | 2001-04-20 | | Release date: | 2001-06-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | The B-Box Dominates Sap-1/Srf Interactions in the Structure of the Ternary Complex
Embo J., 20, 2001
|
|
6J57
 
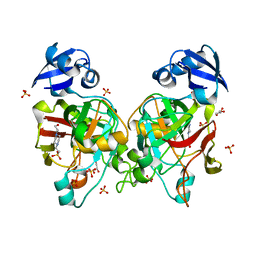 | | Crystal structure of fumarylpyruvate hydrolase from Corynebacterium glutamicum | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, Predicted 2-keto-4-pentenoate hydratase/2-oxohepta-3-ene-1,7-dioic acid hydratase, ... | | Authors: | Hong, H, Seo, H, Kim, K.-J, Park, W. | | Deposit date: | 2019-01-10 | | Release date: | 2019-12-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Sequence, structure and function-based classification of the broadly conserved FAH superfamily reveals two distinct fumarylpyruvate hydrolase subfamilies.
Environ.Microbiol., 22, 2020
|
|
6J5Y
 
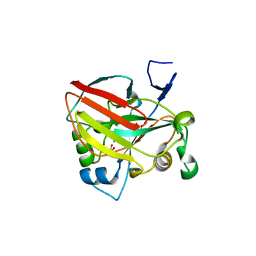 | | Crystal structure of fumarylpyruvate hydrolase from Pseudomonas aeruginosa in complex with Mn2+ and pyruvate | | Descriptor: | FAA hydrolase family protein, MANGANESE (II) ION, PYRUVIC ACID | | Authors: | Hong, H, Seo, H, Kim, K.-J, Park, W. | | Deposit date: | 2019-01-12 | | Release date: | 2019-12-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Sequence, structure and function-based classification of the broadly conserved FAH superfamily reveals two distinct fumarylpyruvate hydrolase subfamilies.
Environ.Microbiol., 22, 2020
|
|
5VL5
 
 | | Coordination Chemistry within a Protein Host: Regulation of the Secondary Coordination Sphere | | Descriptor: | COPPER (II) ION, Streptavidin, [N-(3-{bis[2-(pyridin-2-yl-kappaN)ethyl]amino-kappaN}propyl)-5-(2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl)pentanamide](azido)(hydroxy)copper | | Authors: | Mann, S.I, Heinisch, T, Ward, T.R, Borovik, A.S. | | Deposit date: | 2017-04-24 | | Release date: | 2018-04-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Coordination chemistry within a protein host: regulation of the secondary coordination sphere.
Chem. Commun. (Camb.), 54, 2018
|
|
5VL8
 
 | | Coordination Chemistry within a Protein Host: Regulation of the Secondary Coordination Sphere | | Descriptor: | COPPER (II) ION, GLYCEROL, Streptavidin, ... | | Authors: | Mann, S.I, Heinisch, T, Ward, T.R, Borovik, A.S. | | Deposit date: | 2017-04-25 | | Release date: | 2018-04-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Coordination chemistry within a protein host: regulation of the secondary coordination sphere.
Chem. Commun. (Camb.), 54, 2018
|
|
3LNB
 
 | | Crystal Structure Analysis of Arylamine N-acetyltransferase C from Bacillus anthracis | | Descriptor: | COENZYME A, FORMIC ACID, N-acetyltransferase family protein | | Authors: | Li de la Sierra-Gallay, I, Pluvinage, B, Rodrigues-Lima, F. | | Deposit date: | 2010-02-02 | | Release date: | 2011-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The Bacillus anthracis arylamine N-acetyltransferase ((BACAN)NAT1) that inactivates sulfamethoxazole, reveals unusual structural features compared with the other NAT isoenzymes.
Febs Lett., 585, 2011
|
|
3L8V
 
 | |
5NCO
 
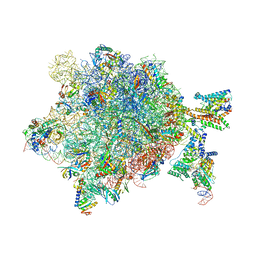 | | Quaternary complex between SRP, SR, and SecYEG bound to the translating ribosome | | Descriptor: | 23S rRNA, 4.5S SRP RNA (Ffs), 50S ribosomal protein L10, ... | | Authors: | Jomaa, A, Hwang Fu, Y, Boerhinger, D, Leibundgut, M, Shan, S.O, Ban, N. | | Deposit date: | 2017-03-06 | | Release date: | 2016-01-27 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structure of the quaternary complex between SRP, SR, and translocon bound to the translating ribosome.
Nat Commun, 8, 2017
|
|
6J5X
 
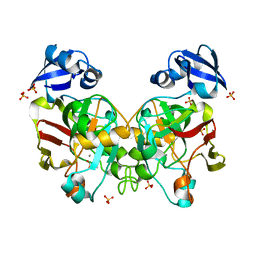 | | Crystal structure of fumarylpyruvate hydrolase from Corynebacterium glutamicum in complex with Mn2+ and pyruvate | | Descriptor: | MANGANESE (II) ION, PYRUVIC ACID, Predicted 2-keto-4-pentenoate hydratase/2-oxohepta-3-ene-1,7-dioic acid hydratase, ... | | Authors: | Hong, H, Seo, H, Kim, K.-J, Park, W. | | Deposit date: | 2019-01-12 | | Release date: | 2019-12-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Sequence, structure and function-based classification of the broadly conserved FAH superfamily reveals two distinct fumarylpyruvate hydrolase subfamilies.
Environ.Microbiol., 22, 2020
|
|
5VKX
 
 | |
4JLQ
 
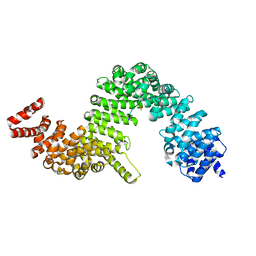 | | Crystal structure of human Karyopherin-beta2 bound to the PY-NLS of Saccharomyces cerevisiae NAB2 | | Descriptor: | Nuclear polyadenylated RNA-binding protein NAB2, Transportin-1 | | Authors: | Sampathkumar, P, Gizzi, A, Rout, M.P, Chook, Y.M, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Nucleocytoplasmic Transport: a Target for Cellular Control (NPCXstals) | | Deposit date: | 2013-03-12 | | Release date: | 2013-04-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Crystal structure of human Karyopherin beta 2 bound to the PY-NLS of Saccharomyces cerevisiae Nab2.
J.Struct.Funct.Genom., 14, 2013
|
|
