5R29
 
 | | PanDDA analysis group deposition -- Endothiapepsin in complex with fragment F2X-Entry G09, DMSO-free | | Descriptor: | (2R)-2-(acetylamino)-4-phenylbutanoic acid, Endothiapepsin | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.105 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5R2R
 
 | | PanDDA analysis group deposition -- Auto-refined data of Endothiapepsin for ground state model 15, DMSO-Free | | Descriptor: | Endothiapepsin | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.048 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5R38
 
 | | PanDDA analysis group deposition -- Auto-refined data of Endothiapepsin for ground state model 32, DMSO-Free | | Descriptor: | Endothiapepsin | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.076 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5R3N
 
 | | PanDDA analysis group deposition -- Auto-refined data of Endothiapepsin for ground state model 47, DMSO-Free | | Descriptor: | Endothiapepsin | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-06-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.059 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5R1V
 
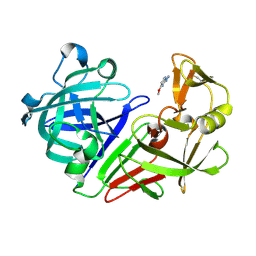 | | PanDDA analysis group deposition -- Endothiapepsin in complex with fragment F2X-Entry C04, DMSO-free | | Descriptor: | 2-hydrazinyl-4-methoxypyrimidine, Endothiapepsin | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5R2A
 
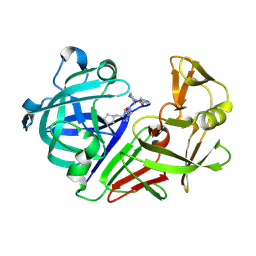 | | PanDDA analysis group deposition -- Endothiapepsin in complex with fragment F2X-Entry H02, DMSO-free | | Descriptor: | 3-cyclopentyl-1-(piperazin-1-yl)propan-1-one, Endothiapepsin | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.049 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5R2N
 
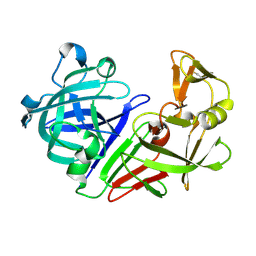 | | PanDDA analysis group deposition -- Auto-refined data of Endothiapepsin for ground state model 11, DMSO-Free | | Descriptor: | Endothiapepsin | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-06-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.078 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5R33
 
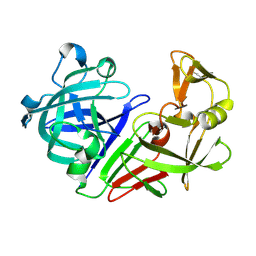 | | PanDDA analysis group deposition -- Auto-refined data of Endothiapepsin for ground state model 27, DMSO-Free | | Descriptor: | Endothiapepsin | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5R3I
 
 | | PanDDA analysis group deposition -- Auto-refined data of Endothiapepsin for ground state model 42, DMSO-Free | | Descriptor: | Endothiapepsin | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.039 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5R3Y
 
 | | PanDDA analysis group deposition -- Auto-refined data of Endothiapepsin for ground state model 58, DMSO-Free | | Descriptor: | Endothiapepsin | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-06-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.038 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
6A8B
 
 | | Ribokinase from Leishmania donovani with AMPPCP | | Descriptor: | CHLORIDE ION, GLYCEROL, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Gatreddi, S, Pillalamarri, V, Qureshi, I.A. | | Deposit date: | 2018-07-06 | | Release date: | 2019-07-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Unraveling structural insights of ribokinase from Leishmania donovani.
Int.J.Biol.Macromol., 136, 2019
|
|
6AMQ
 
 | |
5ZFT
 
 | |
5IQF
 
 | |
5IQI
 
 | |
5ZG6
 
 | |
5ZDO
 
 | | Crystal Structure Analysis of TtQRS in co-crystallised with ATP | | Descriptor: | CHLORIDE ION, Glutamine-tRNA ligase | | Authors: | Mutharasappan, N, Jain, V, Sharma, A, Manickam, Y, Jeyaraman, J. | | Deposit date: | 2018-02-23 | | Release date: | 2018-12-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and functional analysis of Glutaminyl-tRNA synthetase (TtGlnRS) from Thermus thermophilus HB8 and its complexes
Int. J. Biol. Macromol., 120, 2018
|
|
5J7Q
 
 | | Macrophage Migration Inhibitory Factor bound to Inhibitor K664 Derivative | | Descriptor: | 4-(imidazo[1,2-a]pyridin-2-yl)benzene-1,2-diol, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Robertson, M.J, Jorgensen, W.L. | | Deposit date: | 2016-04-06 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Irregularities in enzyme assays: The case of macrophage migration inhibitory factor.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
6B1V
 
 | | Crystal structure of Ps i-CgsB C78S in complex with i-neocarratetraose | | Descriptor: | 1,2-ETHANEDIOL, 3,6-anhydro-2-O-sulfo-alpha-D-galactopyranose-(1-3)-4-O-sulfo-beta-D-galactopyranose-(1-4)-3,6-anhydro-2-O-sulfo-alpha-D-galactopyranose-(1-3)-4-O-sulfo-beta-D-galactopyranose, CALCIUM ION, ... | | Authors: | Hettle, A.G, Boraston, A.B. | | Deposit date: | 2017-09-19 | | Release date: | 2018-03-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | The Molecular Basis of Polysaccharide Sulfatase Activity and a Nomenclature for Catalytic Subsites in this Class of Enzyme.
Structure, 26, 2018
|
|
5ZDL
 
 | | Crystal Structure Analysis of TtQRS in co-crystallised with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, Glutamine--tRNA ligase | | Authors: | Mutharasappan, N, Jain, V, Sharma, A, Manickam, Y, Jeyaraman, J. | | Deposit date: | 2018-02-23 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and functional analysis of Glutaminyl-tRNA synthetase (TtGlnRS) from Thermus thermophilus HB8 and its complexes
Int. J. Biol. Macromol., 120, 2018
|
|
5HBL
 
 | | Native rhodanese domain of YgaP prepared with 1mM DDT is S-nitrosylated | | Descriptor: | Inner membrane protein YgaP | | Authors: | Eichmann, C, Tzitzilonis, C, Nakamura, T, Kwiatkowski, W, Maslennikov, I, Choe, S, Lipton, S.A, Riek, R. | | Deposit date: | 2015-12-31 | | Release date: | 2016-08-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.617 Å) | | Cite: | S-Nitrosylation Induces Structural and Dynamical Changes in a Rhodanese Family Protein.
J.Mol.Biol., 428, 2016
|
|
6A4N
 
 | | HEWL crystals soaked in 2.5M GuHCl for 8 minutes | | Descriptor: | CHLORIDE ION, GLYCEROL, GUANIDINE, ... | | Authors: | Tushar, R, Kini, R.M, Koh, C.Y, Hosur, M.V. | | Deposit date: | 2018-06-20 | | Release date: | 2019-03-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | X-ray crystallographic analysis of time-dependent binding of guanidine hydrochloride to HEWL: First steps during protein unfolding.
Int. J. Biol. Macromol., 122, 2019
|
|
5JMG
 
 | | X-ray structure of the complex between bovine pancreatic ribonuclease and pentachlorocarbonyliridate(III) (4 days of soaking) | | Descriptor: | CARBON MONOXIDE, CHLORIDE ION, IRIDIUM ION, ... | | Authors: | Caterino, M, Petruk, A.A, Vergara, A, Ferraro, G, Merlino, A. | | Deposit date: | 2016-04-29 | | Release date: | 2016-07-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mapping the protein-binding sites for iridium(iii)-based CO-releasing molecules.
Dalton Trans, 45, 2016
|
|
5JML
 
 | | X-ray structure of the complex between bovine pancreatic ribonuclease and penthachlorocarbonyliridate(III) (2 months of soaking) | | Descriptor: | CARBON MONOXIDE, CHLORIDE ION, IRIDIUM ION, ... | | Authors: | Caterino, M, Petruk, A.A, Vergara, A, Ferraro, G, Merlino, A. | | Deposit date: | 2016-04-29 | | Release date: | 2016-07-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Mapping the protein-binding sites for iridium(iii)-based CO-releasing molecules.
Dalton Trans, 45, 2016
|
|
5J16
 
 | |
