7XJH
 
 | | Isoproterenol-activated dog beta3 adrenergic receptor | | Descriptor: | Beta-3 adrenergic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Shihoya, W, Nureki, O. | | Deposit date: | 2022-04-18 | | Release date: | 2022-05-04 | | Last modified: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of the beta 3-adrenergic receptor reveals the molecular basis of subtype selectivity.
Mol.Cell, 81, 2021
|
|
4RLO
 
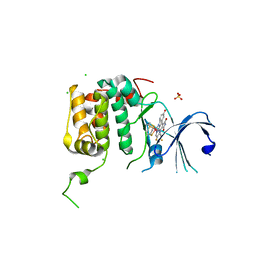 | | Human p70s6k1 with ruthenium-based inhibitor EM5 | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Domsic, J.F, Barber-Rotenberg, J, Salami, J, Qin, J, Marmorstein, R. | | Deposit date: | 2014-10-17 | | Release date: | 2015-01-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.527 Å) | | Cite: | Development of Organometallic S6K1 Inhibitors.
J.Med.Chem., 58, 2015
|
|
3IXJ
 
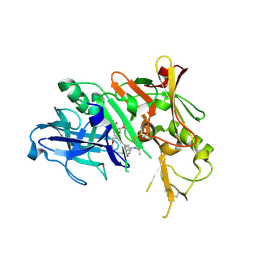 | | Crystal structure of beta-secretase 1 in complex with selective beta-secretase 1 inhibitor | | Descriptor: | Beta-secretase 1, N-[4-(1-BENZYLCARBAMOYL-2-METHYL-PROPYLCARBAMOYL)-1-(3,5-DIFLUORO-PHENOXYMETHYL)-2-HYDROXY-4-METHOXY-BUTYL]-5-(METHANESULFONYL-METHYL-AMINO)-N'-(1-PHENYLETHYL)-ISOPHTHALAMIDE, SULFATE ION | | Authors: | Borkakoti, N, Lindberg, J, Nystrom, S. | | Deposit date: | 2009-09-04 | | Release date: | 2010-03-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design and Synthesis of Potent and Selective BACE-1 Inhibitors.
J.Med.Chem., 53, 2010
|
|
1YCJ
 
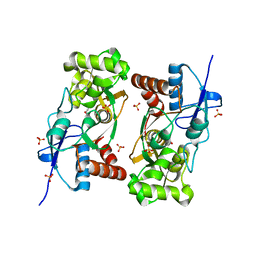 | | Crystal structure of the kainate receptor GluR5 ligand-binding core in complex with (S)-glutamate | | Descriptor: | GLUTAMIC ACID, Ionotropic glutamate receptor 5, SULFATE ION | | Authors: | Naur, P, Vestergaard, B, Skov, L.K, Egebjerg, J, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2004-12-22 | | Release date: | 2005-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the kainate receptor GluR5 ligand-binding core in complex with (S)-glutamate
Febs Lett., 579, 2005
|
|
1MI6
 
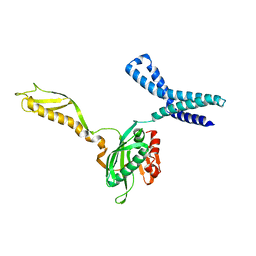 | | Docking of the modified RF2 X-ray structure into the Low Resolution Cryo-EM map of RF2 E.coli 70S Ribosome | | Descriptor: | peptide chain release factor RF-2 | | Authors: | Rawat, U.B.S, Zavialov, A.V, Sengupta, J, Valle, M, Grassucci, R.A, Linde, J, Vestergaard, B, Ehrenberg, M, Frank, J. | | Deposit date: | 2002-08-22 | | Release date: | 2003-01-14 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (12.8 Å) | | Cite: | A cryo-electron microscopic study of ribosome-bound termination factor RF2
Nature, 421, 2003
|
|
7Z34
 
 | | Structure of pre-60S particle bound to DRG1(AFG2). | | Descriptor: | 35S pre-ribosomal RNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Prattes, M, Grishkovskaya, I, Bergler, H, Haselbach, D. | | Deposit date: | 2022-03-01 | | Release date: | 2022-09-21 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Visualizing maturation factor extraction from the nascent ribosome by the AAA-ATPase Drg1.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7Z11
 
 | | Structure of substrate bound DRG1 (AFG2) | | Descriptor: | ATPase family gene 2 protein, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, peptide substrate | | Authors: | Prattes, M, Grishkovskaya, I, Bergler, H, Haselbach, D. | | Deposit date: | 2022-02-24 | | Release date: | 2022-09-21 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Visualizing maturation factor extraction from the nascent ribosome by the AAA-ATPase Drg1.
Nat.Struct.Mol.Biol., 29, 2022
|
|
1MVF
 
 | | MazE addiction antidote | | Descriptor: | PemI-like protein 1, immunoglobulin heavy chain variable region | | Authors: | Loris, R, Marianovsky, I, Lah, J, Laeremans, T, Engelberg-Kulka, H, Glaser, G, Muyldermans, S, Wyns, L. | | Deposit date: | 2002-09-25 | | Release date: | 2003-06-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of the intrinsically flexible addiction antidote MazE.
J.Biol.Chem., 278, 2003
|
|
3CAW
 
 | | Crystal structure of o-succinylbenzoate synthase from Bdellovibrio bacteriovorus liganded with Mg | | Descriptor: | MAGNESIUM ION, o-succinylbenzoate synthase | | Authors: | Fedorov, A.A, Fedorov, E.V, Sakai, A, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-02-20 | | Release date: | 2008-03-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Loss of quaternary structure is associated with rapid sequence divergence in the OSBS family.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3CBS
 
 | | CELLULAR RETINOIC ACID BINDING PROTEIN II IN COMPLEX WITH A SYNTHETIC RETINOIC ACID (RO-12 7310) | | Descriptor: | (2E,4E,6E,8E)-9-(4-hydroxy-2,3,6-trimethylphenyl)-3,7-dimethylnona-2,4,6,8-tetraenoic acid, PROTEIN (CRABP-II) | | Authors: | Chaudhuri, B, Kleywegt, G.J, Bergfors, T, Jones, T.A. | | Deposit date: | 1999-02-22 | | Release date: | 1999-12-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of cellular retinoic acid binding proteins I and II in complex with synthetic retinoids.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
7XQN
 
 | |
3CBA
 
 | |
1YM3
 
 | | Crystal Structure of carbonic anhydrase RV3588c from Mycobacterium tuberculosis | | Descriptor: | CARBONIC ANHYDRASE (CARBONATE DEHYDRATASE) (CARBONIC DEHYDRATASE), MAGNESIUM ION, ZINC ION | | Authors: | Covarrubias, A.S, Larsson, A.M, Hogbom, M, Lindberg, J, Bergfors, T, Bjorkelid, C, Mowbray, S.L, Unge, T, Jones, T.A, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-01-20 | | Release date: | 2005-03-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and function of carbonic anhydrases from Mycobacterium tuberculosis.
J.Biol.Chem., 280, 2005
|
|
7Z2A
 
 | | P. berghei kinesin-8B motor domain in no nucleotide state bound to tubulin dimer | | Descriptor: | Detyrosinated tubulin alpha-1B chain, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-8, ... | | Authors: | Liu, T, Shilliday, F, Cook, A.D, Moores, C.A. | | Deposit date: | 2022-02-26 | | Release date: | 2022-10-19 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Mechanochemical tuning of a kinesin motor essential for malaria parasite transmission.
Nat Commun, 13, 2022
|
|
1ABA
 
 | | THE STRUCTURE OF OXIDIZED BACTERIOPHAGE T4 GLUTAREDOXIN (THIOREDOXIN). REFINEMENT OF NATIVE AND MUTANT PROTEINS | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTAREDOXIN | | Authors: | Eklund, H, Ingelman, M, Soderberg, B.-O, Uhlin, T, Nordlund, P, Nikkola, M, Sonnerstam, U, Joelson, T, Petratos, K. | | Deposit date: | 1992-04-24 | | Release date: | 1993-10-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of oxidized bacteriophage T4 glutaredoxin (thioredoxin). Refinement of native and mutant proteins.
J.Mol.Biol., 228, 1992
|
|
7YX3
 
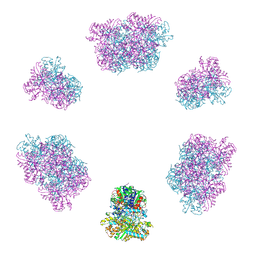 | | Structure of the Mimivirus genomic fibre in its compact 6-start helix form | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative GMC-type oxidoreductase | | Authors: | Villalta, A, Schmitt, A, Estrozi, L.F, Quemin, E.R.J, Alempic, J.M, Lartigue, A, Prazak, V, Belmudes, L, Vasishtan, D, Colmant, A.M.G, Honore, F.A, Coute, Y, Grunewald, K, Abergel, C. | | Deposit date: | 2022-02-15 | | Release date: | 2022-08-10 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | The giant mimivirus 1.2 Mb genome is elegantly organized into a 30-nm diameter helical protein shield.
Elife, 11, 2022
|
|
7YX4
 
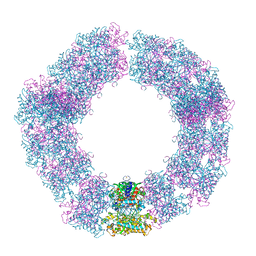 | | Structure of the Mimivirus genomic fibre in its compact 5-start helix form | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative glucose-methanol-choline oxidoreductase protein | | Authors: | Villalta, A, Schmitt, A, Estrozi, L.F, Quemin, E.R.J, Alempic, J.M, Lartigue, A, Prazak, V, Belmudes, L, Vasishtan, D, Colmant, A.M.G, Honore, F.A, Coute, Y, Grunewald, K, Abergel, C. | | Deposit date: | 2022-02-15 | | Release date: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The giant mimivirus 1.2 Mb genome is elegantly organized into a 30 nm diameter helical protein shield.
Elife, 11, 2022
|
|
7YX5
 
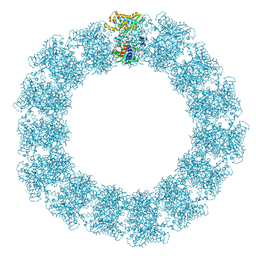 | | Structure of the Mimivirus genomic fibre in its relaxed 5-start helix form | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative glucose-methanol-choline oxidoreductase protein | | Authors: | Villalta, A, Schmitt, A, Estrozi, L.F, Quemin, E.R.J, Alempic, J.M, Lartigue, A, Prazak, V, Belmudes, L, Vasishtan, D, Colmant, A.M.G, Honore, F.A, Coute, Y, Grunewald, K, Abergel, C. | | Deposit date: | 2022-02-15 | | Release date: | 2022-08-10 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The giant mimivirus 1.2 Mb genome is elegantly organized into a 30 nm diameter helical protein shield.
Elife, 11, 2022
|
|
7Z2B
 
 | | P. berghei kinesin-8B motor domain in AMPPNP state bound to tubulin dimer | | Descriptor: | Detyrosinated tubulin alpha-1B chain, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-8, ... | | Authors: | Liu, T, Shilliday, F, Cook, A.D, Moores, C.A. | | Deposit date: | 2022-02-26 | | Release date: | 2022-11-16 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mechanochemical tuning of a kinesin motor essential for malaria parasite transmission.
Nat Commun, 13, 2022
|
|
1N59
 
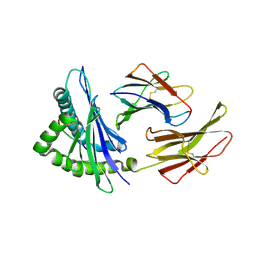 | | Crystal structure of the Murine class I Major Histocompatibility Complex of H-2KB, B2-Microglobulin, and A 9-Residue immunodominant peptide epitope gp33 derived from LCMV | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, K-B alpha chain, ... | | Authors: | Achour, A, Michaelsson, J, Harris, R.A, Odeberg, J, Grufman, P, Sandberg, J.K, Levitsky, V, Karre, K, Sandalova, T, Schneider, G. | | Deposit date: | 2002-11-05 | | Release date: | 2003-01-07 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | A Structural Basis for LCMV Immune Evasion. Subversion of H-2D(b) and H-2K(b) Presentation of gp33
Revealed by Comparative Crystal Structure Analyses.
Immunity, 17, 2002
|
|
1N5A
 
 | | Crystal structure of the Murine class I Major Histocompatibility Complex of H-2DB, B2-Microglobulin, and A 9-Residue immunodominant peptide epitope gp33 derived from LCMV | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, D-B alpha chain, ... | | Authors: | Achour, A, Michaelsson, J, Harris, R.A, Odeberg, J, Grufman, P, Sandberg, J.K, Levitsky, V, Karre, K, Sandalova, T, Schneider, G. | | Deposit date: | 2002-11-05 | | Release date: | 2003-01-07 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A Structural Basis for LCMV Immune Evasion. Subversion of H-2D(b) and H-2K(b) Presentation of
gp33 Revealed by Comparative Crystal Structure Analyses.
Immunity, 17, 2002
|
|
7YYO
 
 | | Cryo-EM structure of an a-carboxysome RuBisCO enzyme at 2.9 A resolution | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, Ribulose bisphosphate carboxylase large chain, ... | | Authors: | Mann, D, Evans, S.L, Bergeron, J.R.C. | | Deposit date: | 2022-02-18 | | Release date: | 2023-01-25 | | Last modified: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Single-particle cryo-EM analysis of the shell architecture and internal organization of an intact alpha-carboxysome.
Structure, 31, 2023
|
|
2GFK
 
 | | Crystal structure of the zinc-beta-lactamase l1 from stenotrophomonas maltophilia (inhibitor 2) | | Descriptor: | 2,5-DIPHENYLFURAN-3,4-DICARBOXYLIC ACID, DI(HYDROXYETHYL)ETHER, Metallo-beta-lactamase L1, ... | | Authors: | Nauton, L, Garau, G, Kahn, R, Dideberg, O. | | Deposit date: | 2006-03-22 | | Release date: | 2007-03-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into the design of inhibitors for the L1 metallo-beta-lactamase from Stenotrophomonas maltophilia.
J.Mol.Biol., 375, 2008
|
|
4NKS
 
 | |
3CS9
 
 | | Human ABL kinase in complex with nilotinib | | Descriptor: | Nilotinib, Proto-oncogene tyrosine-protein kinase ABL1 | | Authors: | Cowan-Jacob, S.W, Fendrich, G, Manley, P, Liebetanz, J, Fabbro, D. | | Deposit date: | 2008-04-09 | | Release date: | 2008-04-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Characterization of AMN107, a selective inhibitor of native and mutant Bcr-Abl
Cancer Cell, 7, 2005
|
|
