6VE9
 
 | |
6V5L
 
 | | The HADDOCK structure model of GDP KRas in complex with its allosteric inhibitor E22 | | Descriptor: | (2R)-2-[2-(1H-indole-3-carbonyl)hydrazinyl]-2-phenylacetamide, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Wang, X, Gupta, A.K, Prakash, P, Putkey, J.P, Gorfe, A.A. | | Deposit date: | 2019-12-04 | | Release date: | 2019-12-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Multi target ensemble based virtual screening yields novel allosteric KRAS inhibitors at high success rate
Chemical Biology & Drug Design, 94, 2019
|
|
8V9U
 
 | |
6QAY
 
 | |
5NQ4
 
 | | Cytotoxin-1 in DPC-micelle | | Descriptor: | Cytotoxin 1 | | Authors: | Dubovskii, P.V, Dubinnyi, M.A, Volynsky, P.E, Pustovalova, Y.E, Konshina, A.G, Utkin, Y.N, Efremov, R.G, Arseniev, A.S. | | Deposit date: | 2017-04-19 | | Release date: | 2017-12-13 | | Last modified: | 2019-05-08 | | Method: | SOLUTION NMR | | Cite: | Impact of membrane partitioning on the spatial structure of an S-type cobra cytotoxin.
J. Biomol. Struct. Dyn., 36, 2018
|
|
7D8K
 
 | | Solution structure of the methyl-CpG binding domain of MBD6 from Arabidopsis thaliana | | Descriptor: | Methyl-CpG-binding domain-containing protein 6 | | Authors: | Mahana, Y, Ohki, I, Walinda, E, Morimoto, D, Sugase, K, Shirakawa, M. | | Deposit date: | 2020-10-08 | | Release date: | 2021-10-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Insights into Methylated DNA Recognition by the Methyl-CpG Binding Domain of MBD6 from Arabidopsis thaliana .
Acs Omega, 7, 2022
|
|
8CX6
 
 | | TPX2 Minimal Active Domain on Microtubules | | Descriptor: | Targeting protein for Xklp2-A | | Authors: | Guo, C, Alfaro-Aco, R, Russell, R, Zhang, C, Petry, S, Polenova, T. | | Deposit date: | 2022-05-19 | | Release date: | 2023-06-28 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Structural basis of protein condensation on microtubules underlying branching microtubule nucleation.
Nat Commun, 14, 2023
|
|
7WKF
 
 | | Antimicrobial peptide-LaIT2 | | Descriptor: | Beta-KTx-like peptide LaIT2 | | Authors: | Tamura, M, Morita, H, Ohki, S. | | Deposit date: | 2022-01-09 | | Release date: | 2023-04-05 | | Method: | SOLUTION NMR | | Cite: | Structural and functional studies of LaIT2, an antimicrobial and insecticidal peptide from Liocheles australasiae.
Toxicon, 214, 2022
|
|
5SYQ
 
 | | Solution structure of Aquifex aeolicus Aq1974 | | Descriptor: | Uncharacterized protein aq_1974 | | Authors: | Sachleben, J.R, Gawlak, G, Hoey, R.J, Liu, G, Joachimiak, A, Montelione, G.T, Koide, S, Northeast Structural Genomics Consortium (NESG), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-08-11 | | Release date: | 2016-09-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Aromatic claw: A new fold with high aromatic content that evades structural prediction.
Protein Sci., 26, 2017
|
|
6X1K
 
 | | Solution NMR structure of de novo designed TMB2.3 | | Descriptor: | De novo designed transmembrane beta-barrel TMB2.3 | | Authors: | Liang, B, Vorobieva, A.A, Chow, C.M, Baker, D, Tamm, L.K. | | Deposit date: | 2020-05-19 | | Release date: | 2021-02-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | De novo design of transmembrane beta barrels.
Science, 371, 2021
|
|
8C5J
 
 | | Spatial structure of Lch-alpha peptide from two-component lantibiotic system Lichenicidin VK21 | | Descriptor: | Lantibiotic lichenicidin VK21 A1 | | Authors: | Mineev, K.S, Paramonov, A.S, Arseniev, A.S, Ovchinnikova, T.V, Shenkarev, Z.O. | | Deposit date: | 2023-01-09 | | Release date: | 2023-03-22 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Specific Binding of the alpha-Component of the Lantibiotic Lichenicidin to the Peptidoglycan Precursor Lipid II Predetermines Its Antimicrobial Activity.
Int J Mol Sci, 24, 2023
|
|
5NHQ
 
 | | Nuclear Magnetic Resonance Structure of the Human Polyoma JC Virus Agnoprotein | | Descriptor: | Agnoprotein | | Authors: | Coric, P, Saribas, A.S, Abou-Gharbia, M, Childers, W, Condra, J, White, M.K, Safak, M, Bouaziz, S. | | Deposit date: | 2017-03-22 | | Release date: | 2017-04-26 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Nuclear Magnetic Resonance Structure of the Human Polyoma JC Virus Agnoprotein.
J. Cell. Biochem., 118, 2017
|
|
5NKO
 
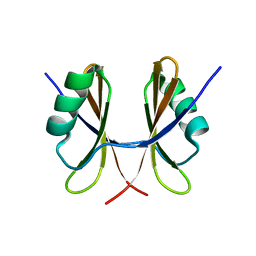 | | Solution structure of the C-terminal domain of S. aureus Hibernating Promoting Factor (CTD-SaHPF) | | Descriptor: | Ribosome hibernation promotion factor | | Authors: | Usachev, K.S, Khusainov, I.S, Ayupov, R.K, Validov, S.Z, Kieffer, B, Yusupov, M.M. | | Deposit date: | 2017-03-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structures and dynamics of hibernating ribosomes from Staphylococcus aureus mediated by intermolecular interactions of HPF.
EMBO J., 36, 2017
|
|
6YGW
 
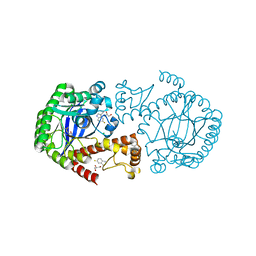 | | TGT W178F mutant labelled mit 5F-Trp crystallised at pH 5.5 | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Nguyen, A, Heine, A, Klebe, G. | | Deposit date: | 2020-03-27 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Mutation study on tRNA-guanine transglycosylase within 19F NMR experiments for conformational change analysis
To Be Published
|
|
6YGP
 
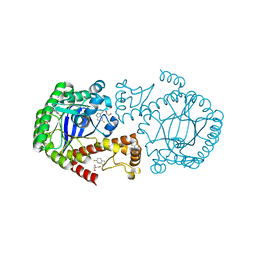 | | TGT WT labelled mit 5F-Trp crystallised at pH 5.5 | | Descriptor: | CHLORIDE ION, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Nguyen, A, Heine, A, Klebe, G. | | Deposit date: | 2020-03-27 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Mutation study on tRNA-guanine transglycosylase within 19F NMR experiments for conformational change analysis
To Be Published
|
|
6YGO
 
 | |
6YGM
 
 | |
6YGL
 
 | |
8C18
 
 | | Solution structure of carotenoid-binding protein AstaPo1 in complex with astaxanthin | | Descriptor: | ASTAXANTHIN, Astaxanthin binding fasciclin family protein | | Authors: | Kornilov, F.D, Savitskaya, A.G, Slonimskiy, Y.B, Goncharuk, S.A, Sluchanko, N.N, Mineev, K.S. | | Deposit date: | 2022-12-20 | | Release date: | 2023-04-05 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the ligand promiscuity of the neofunctionalized, carotenoid-binding fasciclin domain protein AstaP.
Commun Biol, 6, 2023
|
|
6XEH
 
 | |
8DYM
 
 | | Aspartimidylated Graspetide Amycolimiditide | | Descriptor: | ATP-grasp target RiPP | | Authors: | Link, A.J, Choi, B. | | Deposit date: | 2022-08-04 | | Release date: | 2022-11-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Mechanistic Analysis of the Biosynthesis of the Aspartimidylated Graspetide Amycolimiditide.
J.Am.Chem.Soc., 144, 2022
|
|
5NYS
 
 | | M2 G-quadruplex dilute solution | | Descriptor: | DNA (5'-D(*TP*AP*GP*GP*GP*AP*CP*GP*GP*GP*CP*GP*GP*GP*CP*AP*GP*GP*GP*T)-3') | | Authors: | Trajkovski, M, Plavec, J, Endoh, T, Tateishi-Karimata, H, Sugimoto, N. | | Deposit date: | 2017-05-11 | | Release date: | 2018-04-11 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Pursuing origins of (poly)ethylene glycol-induced G-quadruplex structural modulations.
Nucleic Acids Res., 46, 2018
|
|
5NYT
 
 | | M2 G-quadruplex 20 wt% ethylene glycol | | Descriptor: | DNA (5'-D(*TP*AP*GP*GP*GP*AP*CP*GP*GP*GP*CP*GP*GP*GP*CP*AP*GP*GP*GP*T)-3') | | Authors: | Trajkovski, M, Plavec, J, Endoh, T, Tateishi-Karimata, H, Sugimoto, N. | | Deposit date: | 2017-05-11 | | Release date: | 2018-04-11 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Pursuing origins of (poly)ethylene glycol-induced G-quadruplex structural modulations.
Nucleic Acids Res., 46, 2018
|
|
5NYU
 
 | | M2 G-quadruplex 10 wt% PEG8000 | | Descriptor: | DNA (5'-D(*TP*AP*GP*GP*GP*AP*CP*GP*GP*GP*CP*GP*GP*GP*CP*AP*GP*GP*GP*T)-3') | | Authors: | Trajkovski, M, Plavec, J, Endoh, T, Tateishi-Karimata, H, Sugimoto, N. | | Deposit date: | 2017-05-11 | | Release date: | 2018-04-11 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Pursuing origins of (poly)ethylene glycol-induced G-quadruplex structural modulations.
Nucleic Acids Res., 46, 2018
|
|
2YT1
 
 | | Solution structure of the chimera of the C-terminal tail peptide of APP and the C-terminal PID domain of Fe65L | | Descriptor: | Amyloid beta A4 protein and Amyloid beta A4 precursor protein-binding family B member 2 | | Authors: | Li, H, Koshiba, S, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-05 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the C-terminal phosphotyrosine interaction domain of Fe65L1 complexed with the cytoplasmic tail of amyloid precursor protein reveals a novel peptide binding mode
J.Biol.Chem., 283, 2008
|
|
