5O0W
 
 | | Crystal structure of the complex between Nb474 and Trypanosoma congolense fructose-1,6-bisphosphate aldolase | | Descriptor: | Fructose-bisphosphate aldolase, GLYCEROL, Nb474 | | Authors: | Pinto, J, Magez, S, Sterckx, Y. | | Deposit date: | 2017-05-17 | | Release date: | 2017-09-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structural basis for the high specificity of a Trypanosoma congolense immunoassay targeting glycosomal aldolase.
PLoS Negl Trop Dis, 11, 2017
|
|
5QIE
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of HAO1 in complex with Z2856434894 | | Descriptor: | FLAVIN MONONUCLEOTIDE, Hydroxyacid oxidase 1, ~{N},~{N}-dimethyl-4-[(propan-2-ylamino)methyl]aniline | | Authors: | MacKinnon, S, Bezerra, G.A, Krojer, T, Bradley, A.R, Talon, R, Brandao-Neto, J, Douangamath, A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Oppermann, U, Brennan, P.E, Yue, W.W. | | Deposit date: | 2018-05-22 | | Release date: | 2018-07-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QHJ
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of human FAM83B in complex with FMOPL000709a | | Descriptor: | (2S)-1-{[(2H-1,3-benzodioxol-5-yl)methyl]amino}propan-2-ol, 1,2-ETHANEDIOL, IODIDE ION, ... | | Authors: | Pinkas, D.M, Bufton, J.C, Fox, A.E, Talon, R, Krojer, T, Douangamath, A, Collins, P, Zhang, R, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Bullock, A.N. | | Deposit date: | 2018-05-18 | | Release date: | 2018-12-19 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QJU
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT5 in complex with Z906021418 | | Descriptor: | 1,2-ETHANEDIOL, 5-chloro-2-(propan-2-yl)pyrimidine-4-carboxamide, ADP-sugar pyrophosphatase, ... | | Authors: | Dubianok, Y, Collins, P, Krojer, T, Wright, N, Strain-Damerell, C, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, Huber, K, von Delft, F. | | Deposit date: | 2018-10-31 | | Release date: | 2018-12-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QOS
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCP2 (NUDT20) in complex with Z1187701032 | | Descriptor: | (3S)-N-(pyrimidin-2-yl)azepan-3-amine, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Nelson, E.R, Velupillai, S, Talon, R, Collins, P.M, Krojer, T, Wang, D, Brandao-Neto, J, Douangamath, A, Burgess-Brown, N, Arrowsmith, C.H, Bountra, C, Huber, K, von Delft, F. | | Deposit date: | 2019-02-22 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QRC
 
 | | PanDDA analysis group deposition -- Crystal Structure of human ALAS2A in complex with Z31721798 | | Descriptor: | 3-cyclohexyl-1-(morpholin-4-yl)propan-1-one, 5-aminolevulinate synthase, erythroid-specific, ... | | Authors: | Bezerra, G.A, Foster, W, Bailey, H, Shrestha, L, Krojer, T, Talon, R, Brandao-Neto, J, Douangamath, A, Nicola, B.B, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Brennan, P.E, Yue, W.W. | | Deposit date: | 2019-05-22 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QJP
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT5 in complex with Z1578665941 | | Descriptor: | 1,2-ETHANEDIOL, 1-(3-methyl-1,2,4-thiadiazol-5-yl)-1,4-diazepane, ADP-sugar pyrophosphatase, ... | | Authors: | Dubianok, Y, Collins, P, Krojer, T, Wright, N, Strain-Damerell, C, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, Huber, K, von Delft, F. | | Deposit date: | 2018-10-31 | | Release date: | 2018-12-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QJM
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT5 in complex with Z328695024 | | Descriptor: | 1,2-ETHANEDIOL, ADP-sugar pyrophosphatase, CHLORIDE ION, ... | | Authors: | Dubianok, Y, Collins, P, Krojer, T, Wright, N, Strain-Damerell, C, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, Huber, K, von Delft, F. | | Deposit date: | 2018-10-31 | | Release date: | 2018-12-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QRI
 
 | | PanDDA analysis group deposition -- Crystal Structure of human Brachyury in complex with Z2856434826 | | Descriptor: | CADMIUM ION, T-box transcription factor T, [4-(propan-2-yl)piperazin-1-yl](thiophen-2-yl)methanone | | Authors: | Newman, J.A, Gavard, A.E, Fernandez-Cid, A, Sherestha, L, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2019-05-25 | | Release date: | 2019-07-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QI1
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of human PARP14 Macrodomain 3 in complex with FMOPL000474a | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, Poly [ADP-ribose] polymerase 14, ... | | Authors: | Schuller, M, Talon, R, Krojer, T, Brandao-Neto, J, Douangamath, A, Zhang, R, von Delft, F, Schuler, H, Kessler, B, Knapp, S, Bountra, C, Arrowsmith, C.H, Edwards, A, Elkins, J. | | Deposit date: | 2018-05-21 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5RKY
 
 | | PanDDA analysis group deposition -- Crystal Structure of PHIP in complex with Z1796014543 | | Descriptor: | 1-[(2-fluorophenyl)methyl]-N-methylcyclopropane-1-carboxamide, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Biggin, P.C. | | Deposit date: | 2020-06-02 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | PanDDA analysis group deposition of ground-state model
To Be Published
|
|
5SLU
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z1796014543 | | Descriptor: | 1-[(2-fluorophenyl)methyl]-N-methylcyclopropane-1-carboxamide, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SDP
 
 | | PanDDA analysis group deposition -- Crystal Structure of Porphyromonas gingivalis in complex with Z2277255954 | | Descriptor: | 5-fluoro-1-(1-methylcyclopropane-1-sulfonyl)-1,2,3,6-tetrahydropyridine, Asp/Glu-specific dipeptidyl-peptidase, CHLORIDE ION | | Authors: | Tham, C.T, Coker, J.A, Krojer, T, Foster, W.R, Koekemoer, L, Douangamath, A, Talon, R, Fearon, D, von Delft, F, Yue, W.W, Bountra, C, Bezerra, G.A. | | Deposit date: | 2022-01-20 | | Release date: | 2022-02-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.195 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SLD
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z1246465616 | | Descriptor: | (2R)-3-(3,5-dimethyl-1,2-oxazol-4-yl)-N,N,2-trimethylpropanamide, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.581 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QP1
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCP2 (NUDT20) in complex with Z1190363272 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DCP2 (NUDT20), ... | | Authors: | Nelson, E.R, Velupillai, S, Talon, R, Collins, P.M, Krojer, T, Wang, D, Brandao-Neto, J, Douangamath, A, Burgess-Brown, N, Arrowsmith, C.H, Bountra, C, Huber, K, von Delft, F. | | Deposit date: | 2019-02-22 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QGG
 
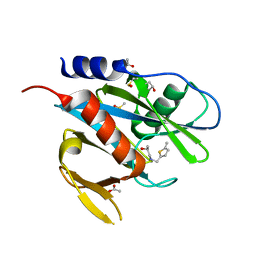 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT7 in complex with FMOPL000693a | | Descriptor: | (2S)-1-{[(5-methylthiophen-2-yl)methyl]amino}propan-2-ol, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Krojer, T, Talon, R, Fairhead, M, Diaz Saez, L, Bradley, A.R, Aimon, A, Collins, P, Brandao-Neto, J, Douangamath, A, Ruda, G.F, Szommer, T, Srikannathasan, V, Elkins, J, Spencer, J, London, N, Nelson, A, Brennan, P.E, Huber, K, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2018-05-15 | | Release date: | 2019-03-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QOR
 
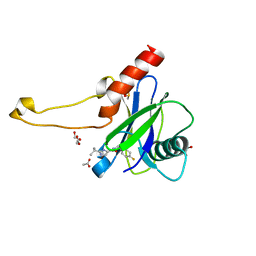 | | PanDDA analysis group deposition -- Crystal Structure of DCP2 (NUDT20) in complex with Z1203490773 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DCP2 (NUDT20), ... | | Authors: | Nelson, E.R, Velupillai, S, Talon, R, Collins, P.M, Krojer, T, Wang, D, Brandao-Neto, J, Douangamath, A, Burgess-Brown, N, Arrowsmith, C.H, Bountra, C, Huber, K, von Delft, F. | | Deposit date: | 2019-02-22 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QP0
 
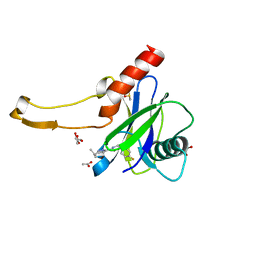 | | PanDDA analysis group deposition -- Crystal Structure of DCP2 (NUDT20) in complex with Z454376544 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DCP2 (NUDT20), ... | | Authors: | Nelson, E.R, Velupillai, S, Talon, R, Collins, P.M, Krojer, T, Wang, D, Brandao-Neto, J, Douangamath, A, Burgess-Brown, N, Arrowsmith, C.H, Bountra, C, Huber, K, von Delft, F. | | Deposit date: | 2019-02-22 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
7H1J
 
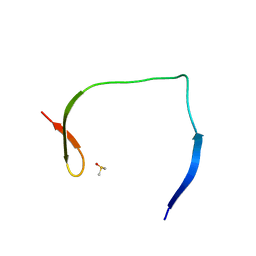 | | PanDDA analysis group deposition -- Crystal Structure of ZIKV NS2B-NS3 protease in complex with Z68299550 | | Descriptor: | 3-chloro-N-(1-hydroxy-2-methylpropan-2-yl)benzamide, DIMETHYL SULFOXIDE, Serine protease NS3, ... | | Authors: | Ni, X, Godoy, A.S, Marples, P.G, Fairhead, M, Balcomb, B.H, Tomlinson, C.W.E, Koekemoer, L, Aschenbrenner, J.C, Lithgo, R.M, Thompson, W, Wild, C, Williams, E.P, Winokan, M, Chandran, A.V, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-04-03 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | PanDDA analysis group deposition of ZIKV NS2B-NS3 protease
To Be Published
|
|
5RH4
 
 | | PanDDA analysis group deposition SARS-CoV-2 main protease fragment screen -- Crystal Structure of SARS-CoV-2 main protease in complex with Z1530425063 (Mpro-x2659) | | Descriptor: | (2R)-2-(6-chloro-9H-carbazol-2-yl)propanoic acid, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-05-15 | | Release date: | 2020-05-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | PanDDA analysis group deposition SARS-CoV-2 main protease fragment screen
To Be Published
|
|
5RH7
 
 | | PanDDA analysis group deposition SARS-CoV-2 main protease fragment screen -- Crystal Structure of SARS-CoV-2 main protease in complex with Z4439011584 (Mpro-x2705) | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-(5-tert-butyl-1H-pyrazol-3-yl)-N-[(1R)-2-[(2-ethyl-6-methylphenyl)amino]-2-oxo-1-(pyridin-3-yl)ethyl]propanamide | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-05-15 | | Release date: | 2020-05-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | PanDDA analysis group deposition SARS-CoV-2 main protease fragment screen
To Be Published
|
|
5RL3
 
 | | PanDDA analysis group deposition of computational designs of SARS-CoV-2 main protease covalent inhibitors -- Crystal Structure of SARS-CoV-2 main protease in complex with LON-WEI-adc59df6-39 (Mpro-x3117) | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-(4-tert-butylphenyl)-N-[(1R)-2-[(oxan-4-yl)amino]-2-oxo-1-(pyridin-3-yl)ethyl]propanamide | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Zaidman, D, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Gorrie-Stone, T.J, Skyner, R, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-08-05 | | Release date: | 2020-12-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | PanDDA analysis group deposition of computational designs of SARS-CoV-2 main protease covalent inhibitors
To Be Published
|
|
5QJO
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT5 in complex with Z57292369 | | Descriptor: | 1,2-ETHANEDIOL, 2-methyl-N-(2-methyl-2H-tetrazol-5-yl)propanamide, ADP-sugar pyrophosphatase, ... | | Authors: | Dubianok, Y, Collins, P, Krojer, T, Wright, N, Strain-Damerell, C, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, Huber, K, von Delft, F. | | Deposit date: | 2018-10-31 | | Release date: | 2018-12-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QI7
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of human PARP14 Macrodomain 3 in complex with FMOPL000506a | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, Poly [ADP-ribose] polymerase 14, ... | | Authors: | Schuller, M, Talon, R, Krojer, T, Brandao-Neto, J, Douangamath, A, Zhang, R, von Delft, F, Schuler, H, Kessler, B, Knapp, S, Bountra, C, Arrowsmith, C.H, Edwards, A, Elkins, J. | | Deposit date: | 2018-05-21 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QJ7
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT5 in complex with Z32327641 | | Descriptor: | 1,2-ETHANEDIOL, ADP-sugar pyrophosphatase, CHLORIDE ION, ... | | Authors: | Dubianok, Y, Collins, P, Krojer, T, Wright, N, Strain-Damerell, C, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, Huber, K, von Delft, F. | | Deposit date: | 2018-10-31 | | Release date: | 2018-12-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
