1OB9
 
 | | Holliday Junction Resolving Enzyme | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, HOLLIDAY JUNCTION RESOLVASE | | Authors: | Middleton, C.L, Parker, J.L, Richard, D.J, White, M.F, Bond, C.S. | | Deposit date: | 2003-01-28 | | Release date: | 2004-10-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substrate Recognition and Catalysis by the Holliday Junction Resolving Enzyme Hje.
Nucleic Acids Res., 32, 2004
|
|
7X3P
 
 | | Crystal structure of human SIRT5 in complex with diazirine inhibitor 9 | | Descriptor: | 5-[[(5~{S})-6-[[(1~{S})-1-(4-hydroxyphenyl)-2-oxidanylidene-2-(prop-2-ynylamino)ethyl]amino]-6-oxidanylidene-5-[[4-[3-(trifluoromethyl)-1,2-diazirin-3-yl]phenyl]carbonylamino]hexyl]amino]-5-sulfanylidene-pentanoic acid, DI(HYDROXYETHYL)ETHER, NAD-dependent protein deacylase sirtuin-5, ... | | Authors: | Li, G.-B, Deng, J. | | Deposit date: | 2022-03-01 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Crystal structure of human SIRT5 in complex with diazirine inhibitor 9
To Be Published
|
|
1E3U
 
 | | MAD structure of OXA10 class D beta-lactamase | | Descriptor: | 1,2-ETHANEDIOL, BETA-LACTAMASE OXA-10, GOLD (I) CYANIDE ION, ... | | Authors: | Maveyraud, L, Golemi, D, Kotra, L.P, Tranier, S, Vakulenko, S, Mobashery, S, Samama, J.P. | | Deposit date: | 2000-06-23 | | Release date: | 2001-01-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Insights Into Class D Beta-Lactamases are Revealed by the Crystal Structure of the Oxa10 Enzyme from Pseudomonas Aeruginosa
Structure, 8, 2000
|
|
4OOX
 
 | |
5E9C
 
 | | Crystal structure of human heparanase in complex with heparin tetrasaccharide dp4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-deoxy-2-O-sulfo-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Wu, L, Davies, G.J. | | Deposit date: | 2015-10-15 | | Release date: | 2015-11-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural characterization of human heparanase reveals insights into substrate recognition.
Nat.Struct.Mol.Biol., 22, 2015
|
|
1I48
 
 | | CYSTATHIONINE GAMMA-SYNTHASE IN COMPLEX WITH THE INHIBITOR CTCPO | | Descriptor: | CARBOXYMETHYLTHIO-3-(3-CHLOROPHENYL)-1,2,4-OXADIAZOL, CYSTATHIONINE GAMMA-SYNTHASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Steegborn, C, Laber, B, Messerschmidt, A, Huber, R, Clausen, T. | | Deposit date: | 2001-02-20 | | Release date: | 2001-09-05 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Crystal structures of cystathionine gamma-synthase inhibitor complexes rationalize the increased affinity of a novel inhibitor.
J.Mol.Biol., 311, 2001
|
|
2VMF
 
 | | Structural and biochemical evidence for a boat-like transition state in beta-mannosidases | | Descriptor: | (5R,6R,7S,8R)-5-(HYDROXYMETHYL)-5,6,7,8-TETRAHYDROIMIDAZO[1,2-A]PYRIDINE-6,7,8-TRIOL, 1,2-ETHANEDIOL, BETA-MANNOSIDASE, ... | | Authors: | Tailford, L.E, Offen, W.A, Smith, N.L, Dumon, C, Moreland, C, Gratien, J, Heck, M.P, Stick, R.V, Bleriot, Y, Vasella, A, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2008-01-25 | | Release date: | 2008-04-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Biochemical Evidence for a Boat-Like Transition State in Beta-Mannosidases.
Nat.Chem.Biol., 4, 2008
|
|
3ZQI
 
 | | Structure of Tetracycline repressor in complex with inducer peptide- TIP2 | | Descriptor: | 1,2-ETHANEDIOL, INDUCER PEPTIDE TIP2, MAGNESIUM ION, ... | | Authors: | Sevvana, M, Goeke, D, Stoeckle, C, Kaspar, D, Grubmueller, S, Goetz, C, Wimmer, C, Berens, C, Klotzsche, M, Muller, Y.A, Hillen, W. | | Deposit date: | 2011-06-09 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | An Exclusive Alpha/Beta Code Directs Allostery in Tetr-Peptide Complexes.
J.Mol.Biol., 416, 2012
|
|
6QFU
 
 | | Human carbonic anhydrase II with bound IrCp* complex (cofactor 7) to generate an artificial transfer hydrogenase (ATHase) | | Descriptor: | 4-[2-(9-chloranyl-2',3',4',5',6'-pentamethyl-4-oxidanyl-7-oxidanylidene-spiro[1$l^{4},8-diaza-9$l^{8}-iridabicyclo[4.3.0]nona-1(6),2,4-triene-9,1'-1$l^{8}-iridapentacyclo[2.2.0.0^{1,3}.0^{1,5}.0^{2,6}]hexane]-8-yl)ethyl]benzenesulfonamide, Carbonic anhydrase 2, SULFATE ION, ... | | Authors: | Rebelein, J.G. | | Deposit date: | 2019-01-10 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Chemical Optimization of Whole-Cell Transfer Hydrogenation Using Carbonic Anhydrase as Host Protein.
Acs Catalysis, 9, 2019
|
|
6QFX
 
 | | Human carbonic anhydrase II with bound IrCp* complex (cofactor 10) to generate an artificial transfer hydrogenase (ATHase) | | Descriptor: | 2-(9-chloranyl-2',3',4',5',6'-pentamethyl-4-oxidanyl-7-oxidanylidene-spiro[1$l^{4},8-diaza-9$l^{8}-iridabicyclo[4.3.0]nona-1,3,5-triene-9,1'-1$l^{8}-iridapentacyclo[2.2.0.0^{1,3}.0^{1,5}.0^{2,6}]hexane]-8-yl)-~{N}-(4-sulfamoylphenyl)ethanamide, Carbonic anhydrase 2, SULFATE ION, ... | | Authors: | Rebelein, J.G. | | Deposit date: | 2019-01-10 | | Release date: | 2019-04-17 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Chemical Optimization of Whole-Cell Transfer Hydrogenation Using Carbonic Anhydrase as Host Protein.
Acs Catalysis, 9, 2019
|
|
5O4E
 
 | | Crystal structure of VEGF in complex with heterodimeric Fcab JanusCT6 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Mlynek, G, Lobner, E, Kubinger, K, Humm, A, Obinger, C, Djinovic-Carugo, K. | | Deposit date: | 2017-05-29 | | Release date: | 2017-08-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Two-faced Fcab prevents polymerization with VEGF and reveals thermodynamics and the 2.15 angstrom crystal structure of the complex.
MAbs, 9, 2017
|
|
5V7W
 
 | | Crystal structure of human PARP14 bound to 2-{[(1-methylpiperidin-4-yl)methyl]amino}-5,6,7,8-tetrahydro[1]benzothieno[2,3-d]pyrimidin-4(3H)-one inhibitor | | Descriptor: | 2-{[(1-methylpiperidin-4-yl)methyl]amino}-5,6,7,8-tetrahydro[1]benzothieno[2,3-d]pyrimidin-4(3H)-one, Poly [ADP-ribose] polymerase 14 | | Authors: | saikatendu, k.s, Hirozane, M. | | Deposit date: | 2017-03-20 | | Release date: | 2017-05-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Identification of PARP14 inhibitors using novel methods for detecting auto-ribosylation.
Biochem. Biophys. Res. Commun., 486, 2017
|
|
3AL9
 
 | | Mouse Plexin A2 extracellular domain | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Plexin-A2 | | Authors: | Nogi, T, Yasui, N, Mihara, E, Takagi, J. | | Deposit date: | 2010-07-28 | | Release date: | 2010-10-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for semaphorin signalling through the plexin receptor.
Nature, 467, 2010
|
|
3ZPI
 
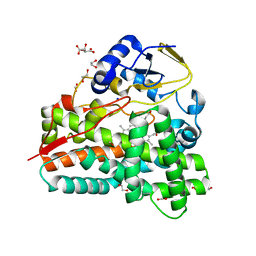 | | PikC D50N mutant in P21 space group | | Descriptor: | 1,2-ETHANEDIOL, CYTOCHROME P450 HYDROXYLASE PIKC, FORMIC ACID, ... | | Authors: | Podust, L.M. | | Deposit date: | 2013-02-27 | | Release date: | 2014-03-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Recognition of Synthetic Substrates by P450 Pikc
To be Published
|
|
4KXI
 
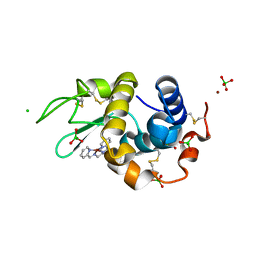 | | Crystallographic study of the complex of Ni(II) Schiff base complex and HEW Lysozyme | | Descriptor: | (N1E, N2E)-N1, N2-bis(pyridine-2-ylmethylene)propane-1,2-diamine, ... | | Authors: | Koley Seth, B, Ray, A, Basu, S, Biswas, S. | | Deposit date: | 2013-05-27 | | Release date: | 2014-05-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic study of the complex of Ni(II) Schiff base complex and HEW Lysozyme
To be Published
|
|
2VI2
 
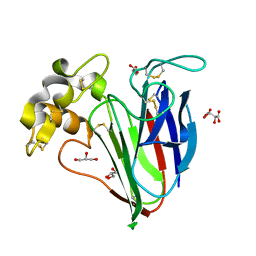 | |
7JRL
 
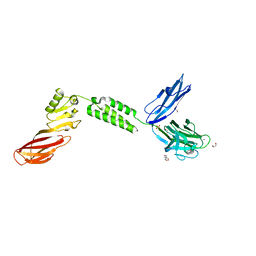 | | The structure of CBM51-2 in complex with GlcNAc and INT domains from Clostridium perfringens ZmpB | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pluvinage, B, Boraston, A.B. | | Deposit date: | 2020-08-12 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Architecturally complex O -glycopeptidases are customized for mucin recognition and hydrolysis.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
2VLY
 
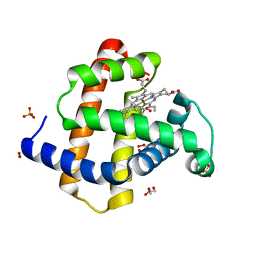 | | Crystal structure of myoglobin compound III (radiation-induced) | | Descriptor: | GLYCEROL, HYDROGEN PEROXIDE, MYOGLOBIN, ... | | Authors: | Hersleth, H.-P, Gorbitz, C.H, Andersson, K.K. | | Deposit date: | 2008-01-20 | | Release date: | 2008-01-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Crystal Structure of Peroxymyoglobin Generated Through Cryoradiolytic Reduction of Myoglobin Compound III During Data Collection.
Biochem.J., 412, 2008
|
|
8T3D
 
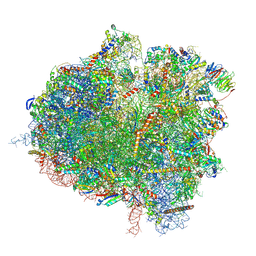 | | Hypomethylated yeast 80S bound with Taura syndrome virus (TSV) internal ribosome entry site (IRES), eEF2, GDP, and sordarin, Structure III | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0-A, ... | | Authors: | Zhao, Y, Li, H. | | Deposit date: | 2023-06-07 | | Release date: | 2024-12-11 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Regulation of Translation by Ribosome RNA 2'-O-Methylation
To be Published
|
|
9MRD
 
 | |
8T3C
 
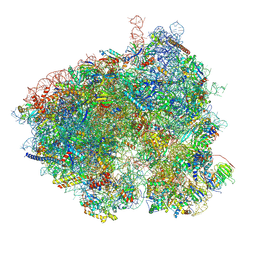 | | Hypomethylated yeast 80S bound with Taura syndrome virus (TSV) internal ribosome entry site (IRES), eEF2, GDP, and sordarin, Structure II | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0-A, ... | | Authors: | Zhao, Y, Li, H. | | Deposit date: | 2023-06-07 | | Release date: | 2024-12-11 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Regulation of Translation by Ribosome RNA 2'-O-Methylation
To Be Published
|
|
3EZ1
 
 | |
1LQD
 
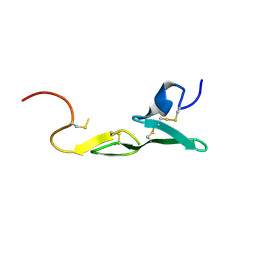 | | CRYSTAL STRUCTURE OF FXA IN COMPLEX WITH 45. | | Descriptor: | 1-(3-CARBAMIMIDOYL-BENZYL)-4-METHYL-1H-INDOLE-2-CARBOXYLIC ACID 3,5-DIMETHYL-BENZYLAMIDE, Blood coagulation factor Xa, CALCIUM ION | | Authors: | Schreuder, H.A, Loenze, P, Brachvogel, V, Liesum, A. | | Deposit date: | 2002-05-10 | | Release date: | 2003-05-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Design and Quantitative Structure-Activity relationship of 3-amidinobenzyl-1H-indole-2-carboxamides as potent, nonchiral, and selective inhibitors of blood coagulation factor Xa
J.Med.Chem., 45, 2002
|
|
2VLZ
 
 | | Crystal structure of peroxymyoglobin generated by cryoradiolytic reduction of myoglobin compound III | | Descriptor: | GLYCEROL, HYDROGEN PEROXIDE, MYOGLOBIN, ... | | Authors: | Hersleth, H.-P, Gorbitz, C.H, Andersson, K.K. | | Deposit date: | 2008-01-20 | | Release date: | 2008-01-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Crystal Structure of Peroxymyoglobin Generated Through Cryoradiolytic Reduction of Myoglobin Compound III During Data Collection.
Biochem.J., 412, 2008
|
|
3RBM
 
 | | Crystal structure of Plasmodium vivax geranylgeranylpyrophosphate synthase complexed with BPH -703 | | Descriptor: | 3-(2,2-diphosphonoethyl)-1-dodecyl-1H-imidazol-3-ium, Farnesyl pyrophosphate synthase, MAGNESIUM ION, ... | | Authors: | Liu, Y.L, No, J.H, Oldfield, E. | | Deposit date: | 2011-03-29 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Lipophilic analogs of zoledronate and risedronate inhibit Plasmodium geranylgeranyl diphosphate synthase (GGPPS) and exhibit potent antimalarial activity.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
