7RDW
 
 | | Crystal Structure of FH1 Fab bound to HXb2 HIV-1 gp120 core | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, AMMONIUM ION, ... | | Authors: | Weidle, C, Pancera, M. | | Deposit date: | 2021-07-12 | | Release date: | 2022-05-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | Characterization of a vaccine-elicited human antibody with sequence homology to VRC01-class antibodies that binds the C1C2 gp120 domain.
Sci Adv, 8, 2022
|
|
3UQ0
 
 | | Crystal structure of the post-catalytic product complex of polymerase lambda with an rAMP at the primer terminus. | | Descriptor: | 1,2-ETHANEDIOL, 5'-D(*CP*AP*GP*TP*AP*CP)-R(P*A)-3', 5'-D(*CP*GP*GP*CP*TP*GP*TP*AP*CP*TP*G)-3', ... | | Authors: | Gosavi, R.A, Moon, A.F, Kunkel, T.A, Pedersen, L.C, Bebenek, K. | | Deposit date: | 2011-11-18 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | The catalytic cycle for ribonucleotide incorporation by human DNA Pol lambda
Nucleic Acids Res., 40, 2012
|
|
3USM
 
 | | Crystal Structure of LeuT bound to L-selenomethionine in space group C2 from lipid bicelles (collected at 1.2 A) | | Descriptor: | IODIDE ION, PHOSPHOCHOLINE, SELENOMETHIONINE, ... | | Authors: | Wang, H, Elferich, J, Gouaux, E. | | Deposit date: | 2011-11-23 | | Release date: | 2012-01-11 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.008 Å) | | Cite: | Structures of LeuT in bicelles define conformation and substrate binding in a membrane-like context.
Nat.Struct.Mol.Biol., 19, 2012
|
|
5XLJ
 
 | | Crystal structure of the flagellar cap protein flid D2-D3 domains from serratia marcescens in Space group P432 | | Descriptor: | CHLORIDE ION, Flagellar hook-associated protein 2, SODIUM ION | | Authors: | Cho, S.Y, Song, W.S, Hong, H.J, Yoon, S.I. | | Deposit date: | 2017-05-10 | | Release date: | 2017-06-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Tetrameric structure of the flagellar cap protein FliD from Serratia marcescens.
Biochem. Biophys. Res. Commun., 489, 2017
|
|
6WRL
 
 | | The interaction of chlorido(1,5-cyclooctadiene)([4-(2-((tert-butoxycarbonyl)amino)-3-methoxy-3-oxopropyl)-1,3-dimethyl-1H-imidazol-3-ide])rhodium(I) with HEWL after 1 week | | Descriptor: | ACETATE ION, Lysozyme, Rhodium, ... | | Authors: | Sullivan, M.P, John, M, Hartinger, C.G, Goldstone, D.C. | | Deposit date: | 2020-04-29 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A Combined Spectroscopic and Protein Crystallography Study Reveals Protein Interactions of Rh I (NHC) Complexes at the Molecular Level.
Inorg.Chem., 59, 2020
|
|
5RBZ
 
 | | PanDDA analysis group deposition -- Endothiapepsin changed state model for fragment F2X-Entry Library E05a | | Descriptor: | 2-(2-methylphenyl)acetamide, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-03-24 | | Release date: | 2020-06-03 | | Last modified: | 2020-06-17 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
6WRN
 
 | | Crystal structure of Mj 3-nitro-tyrosine tRNA synthetase (5B) C70A variant bound to 3-nitro-tyrosine | | Descriptor: | META-NITRO-TYROSINE, SODIUM ION, Tyrosine--tRNA ligase | | Authors: | Beyer, J.N, Hosseinzadeh, P, Karplus, P.A, Mehl, R.A, Cooley, R.B. | | Deposit date: | 2020-04-29 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Overcoming Near-Cognate Suppression in a Release Factor 1-Deficient Host with an Improved Nitro-Tyrosine tRNA Synthetase.
J.Mol.Biol., 432, 2020
|
|
5RCW
 
 | | PanDDA analysis group deposition -- Endothiapepsin ground state model 18 | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, Endothiapepsin, ... | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-03-24 | | Release date: | 2020-06-03 | | Last modified: | 2020-06-17 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
6WUR
 
 | |
7R6T
 
 | | Human EXOG complexed with dRP-containing DNA | | Descriptor: | DNA (5'-D(*CP*GP*CP*AP*CP*GP*TP*CP*AP*GP*A)-3'), DNA (5'-D(3DR)P*CP*TP*GP*AP*CP*GP*TP*GP*C)-3'), MAGNESIUM ION, ... | | Authors: | Szymanski, M.R, Yin, Y.W. | | Deposit date: | 2021-06-23 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Human EXOG possesses strong AP hydrolysis activity
To Be Published
|
|
5XA7
 
 | | Complete structure factors and an atomic model of the calcium pump (SERCA1A) and associated phospholipids in the E1-2CA2+ crystals | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, SODIUM ION, ... | | Authors: | Norimatsu, Y, Hasegawa, K, Shimizu, N, Toyoshima, C. | | Deposit date: | 2017-03-11 | | Release date: | 2017-05-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Protein-phospholipid interplay revealed with crystals of a calcium pump.
Nature, 545, 2017
|
|
7MCN
 
 | | Crystal structure of Staphylococcus aureus Cystathionine gamma-lyase, Holoenzyme with High HEPES | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Bifunctional cystathionine gamma-lyase/homocysteine desulfhydrase, GLYCEROL, ... | | Authors: | Nuthanakanti, A, Serganov, A, Kaushik, A. | | Deposit date: | 2021-04-02 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Inhibitors of bacterial H 2 S biogenesis targeting antibiotic resistance and tolerance.
Science, 372, 2021
|
|
3USZ
 
 | | Crystal structure of truncated exo-1,3/1,4-beta-glucanase (EXOP) from Pseudoalteromonas sp. BB1 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Exo-1,3/1,4-beta-glucanase, ... | | Authors: | Nakatani, Y, Cutfield, S.M, Cutfield, J.F. | | Deposit date: | 2011-11-24 | | Release date: | 2011-12-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and activity of exo-1,3/1,4-beta-glucanase from marine bacterium Pseudoalteromonas sp. BB1 showing a novel C-terminal domain
Febs J., 2011
|
|
6WRM
 
 | | The interaction of chlorido(1,5-cyclooctadiene)([4-(2-((tert-butoxycarbonyl)amino)-3-methoxy-3-oxopropyl)-1,3-dimethyl-1H-imidazol-3-ide])rhodium(I) with HEWL after 1 month | | Descriptor: | (4-{2-[(tert-butoxycarbonyl)amino]-3-methoxy-3-oxopropyl}-1,3-dimethyl-2,3-dihydro-1H-imidazol-2-yl)rhodium, CHLORIDE ION, Lysozyme, ... | | Authors: | Sullivan, M.P, John, M, Hartinger, C.G, Goldstone, D.C. | | Deposit date: | 2020-04-29 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A Combined Spectroscopic and Protein Crystallography Study Reveals Protein Interactions of Rh I (NHC) Complexes at the Molecular Level.
Inorg.Chem., 59, 2020
|
|
6WRQ
 
 | | Crystal structure of Mj 3-nitro-tyrosine tRNA synthetase (5B) S158C variant bound to 3-nitro-tyrosine | | Descriptor: | GLYCEROL, META-NITRO-TYROSINE, SODIUM ION, ... | | Authors: | Beyer, J.N, Hosseinzadeh, P, Karplus, P.A, Mehl, R.A, Cooley, R.B. | | Deposit date: | 2020-04-29 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Overcoming Near-Cognate Suppression in a Release Factor 1-Deficient Host with an Improved Nitro-Tyrosine tRNA Synthetase.
J.Mol.Biol., 432, 2020
|
|
5XH9
 
 | | Aspergillus kawachii beta-fructofuranosidase | | Descriptor: | Extracellular invertase, SODIUM ION | | Authors: | Nagaya, M, Tonozuka, T. | | Deposit date: | 2017-04-19 | | Release date: | 2017-07-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a beta-fructofuranosidase with high transfructosylation activity from Aspergillus kawachii
Biosci. Biotechnol. Biochem., 81, 2017
|
|
3V6O
 
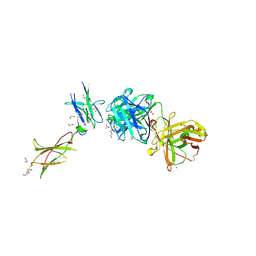 | | Leptin Receptor-antibody complex | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Carpenter, B, Hemsworth, G.R, Ross, R.J, Artymiuk, P.J. | | Deposit date: | 2011-12-20 | | Release date: | 2012-03-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of the human obesity receptor leptin-binding domain reveals the mechanism of leptin antagonism by a monoclonal antibody.
Structure, 20, 2012
|
|
5RBP
 
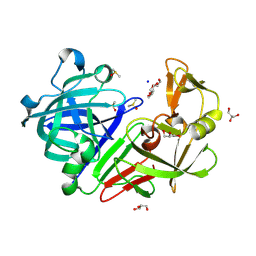 | | PanDDA analysis group deposition -- Endothiapepsin changed state model for fragment F2X-Entry Library B03a | | Descriptor: | 2-[(1~{S})-1-azanylpropyl]phenol, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-03-24 | | Release date: | 2020-06-03 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5RCI
 
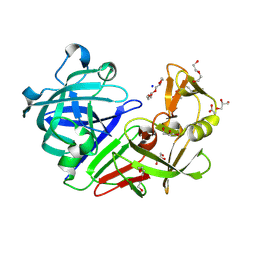 | | PanDDA analysis group deposition -- Endothiapepsin ground state model 03 | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, Endothiapepsin, ... | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-03-24 | | Release date: | 2020-06-03 | | Last modified: | 2020-06-17 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
3VD9
 
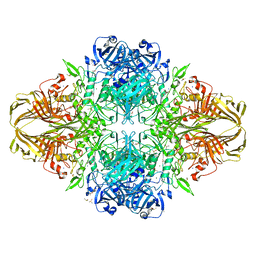 | | E. coli (lacZ) beta-galactosidase (N460S) in complex with IPTG | | Descriptor: | 1-methylethyl 1-thio-beta-D-galactopyranoside, Beta-galactosidase, DIMETHYL SULFOXIDE, ... | | Authors: | Wheatley, R.W, Kappelhoff, J.C, Hahn, J.N, Dugdale, M.L, Dutkoski, M.J, Tamman, S.D, Fraser, M.E, Huber, R.E. | | Deposit date: | 2012-01-04 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Substitution for Asn460 cripples {beta}-galactosidase (Escherichia coli) by increasing substrate affinity and decreasing transition state stability.
Arch.Biochem.Biophys., 521, 2012
|
|
5RD8
 
 | | PanDDA analysis group deposition -- Endothiapepsin ground state model 30 | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, Endothiapepsin, ... | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-03-24 | | Release date: | 2020-06-03 | | Last modified: | 2020-06-17 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
3VDC
 
 | | E. coli (lacZ) beta-galactosidase (N460T) in complex with IPTG | | Descriptor: | 1-methylethyl 1-thio-beta-D-galactopyranoside, Beta-galactosidase, DIMETHYL SULFOXIDE, ... | | Authors: | Wheatley, R.W, Kappelhoff, J.C, Hahn, J.N, Dugdale, M.L, Dutkoski, M.J, Tamman, S.D, Fraser, M.E, Huber, R.E. | | Deposit date: | 2012-01-04 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Substitution for Asn460 cripples {beta}-galactosidase (Escherichia coli) by increasing substrate affinity and decreasing transition state stability.
Arch.Biochem.Biophys., 521, 2012
|
|
6WX5
 
 | | Adducts formed after 3 weeks in the reaction of chlorido[chlorido(2,2'-((2-([2,2':6',2''-Terpyridin]-4'-yloxy)ethyl)azanediyl)bis(ethan-1-ol))platinum(II)] with HEWL | | Descriptor: | ACETATE ION, Lysozyme, PLATINUM (II) ION, ... | | Authors: | Sullivan, M.P, Hartinger, C.G, Goldstone, D.C. | | Deposit date: | 2020-05-09 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Mustards-Derived Terpyridine-Platinum Complexes as Anticancer Agents: DNA Alkylation vs Coordination.
Inorg.Chem., 60, 2021
|
|
7M8Q
 
 | | Complex structure of Methane monooxygenase hydroxylase and regulatory subunit with fluorosubstituted tryptophans | | Descriptor: | 1,2-ETHANEDIOL, BENZOIC ACID, FE (III) ION, ... | | Authors: | Johns, J.C, Banerjee, R, Shi, K, Semonis, M.M, Aihara, H, Pomerantz, W.C.K, Lipscomb, J.D. | | Deposit date: | 2021-03-30 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Soluble Methane Monooxygenase Component Interactions Monitored by 19 F NMR.
Biochemistry, 60, 2021
|
|
5X62
 
 | |
