2XV3
 
 | | Pseudomonas aeruginosa Azurin with mutated metal-binding loop sequence (CAAAAHAAAAM), chemically reduced, pH5.3 | | Descriptor: | AZURIN, COPPER (I) ION | | Authors: | Li, C, Sato, K, Monari, S, Salard, I, Sola, M, Banfield, M.J, Dennison, C. | | Deposit date: | 2010-10-22 | | Release date: | 2010-12-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Metal-Binding Loop Length is a Determinant of the Pka of a Histidine Ligand at a Type 1 Copper Site
Inorg.Chem., 50, 2011
|
|
5CWX
 
 | | Structure of Xoo1075, a peptide deformylase from Xanthomonas oryzae pv oryzae, in complex with fragment 134 | | Descriptor: | 3-(2-methyl-1,3-thiazol-4-yl)aniline, ACETATE ION, CADMIUM ION, ... | | Authors: | Ngo, H.P.T, Kang, L.W. | | Deposit date: | 2015-07-28 | | Release date: | 2016-08-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Xoo1075, a peptide deformylase from Xanthomonas oryzae pv oryzae, in complex with fragment 134
To Be Published
|
|
4D3W
 
 | | Crystal Structure of Epithelial Adhesin 1 A domain (Epa1A) from Candida glabrata in complex with the T-antigen (Galb1-3GalNAc) | | Descriptor: | CALCIUM ION, EPITHELIAL ADHESIN 1, GLYCEROL, ... | | Authors: | Kock, M, Maestre-Reyna, M, Diderrich, R, Moesch, H.-U, Essen, L.-O. | | Deposit date: | 2014-10-24 | | Release date: | 2015-07-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Hotspots Determine Functional Diversity of the Candida Glabrata Epithelial Adhesin Family
J.Biol.Chem., 290, 2015
|
|
4OIF
 
 | | 3D structure of Gan42B, a GH42 beta-galactosidase from G. | | Descriptor: | Beta-galactosidase, GLYCEROL, ZINC ION | | Authors: | Solomon, H.V, Tabachnikov, O, Feinberg, H, Shoham, Y, Shoham, G. | | Deposit date: | 2014-01-19 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.448 Å) | | Cite: | Crystallization and preliminary crystallographic analysis of GanB, a GH42 intracellular beta-galactosidase from Geobacillus stearothermophilus.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
5CYN
 
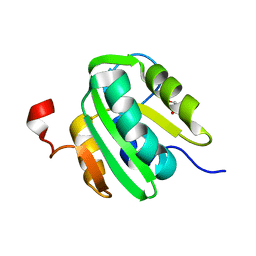 | | JC Virus large T-antigen origin binding domain F258L mutant | | Descriptor: | 1,2-ETHANEDIOL, Large T antigen | | Authors: | Meinke, G, Bohm, A, Bullock, P.A. | | Deposit date: | 2015-07-30 | | Release date: | 2015-12-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Based Analyses of the JC Virus T-Antigen F258L Mutant Provides Evidence for DNA Dependent Conformational Changes in the C-Termini of Polyomavirus Origin Binding Domains.
Plos Pathog., 12, 2016
|
|
3A2J
 
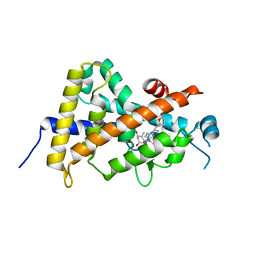 | | Crystal structure of the human vitamin D receptor (H305F/H397F) ligand binding domain complexed with TEI-9647 | | Descriptor: | (1S,3R,5Z,7E,20S,23S)-1,3-dihydroxy-23,26-epoxy-9,10-secocholesta-5,7,10,25(27)-tetraen-26-one, Vitamin D3 receptor | | Authors: | Kakuda, S, Takimoto-Kamimura, M. | | Deposit date: | 2009-05-20 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of the histidine-mediated vitamin D receptor agonistic and antagonistic mechanisms of (23S)-25-dehydro-1alpha-hydroxyvitamin D(3)-26,23-lactone
Acta Crystallogr.,Sect.D, 66, 2010
|
|
2Q3G
 
 | | Structure of the PDZ domain of human PDLIM7 bound to a C-terminal extension from human beta-tropomyosin | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PDZ and LIM domain protein 7 | | Authors: | Gileadi, C, Papagrigoriou, E, Elkins, J, Burgess-Brown, N, Salah, E, Gileadi, O, Umeano, C, Bunkoczi, G, von Delft, F, Uppenberg, J, Pike, A.C.W, Arrowsmith, C.H, Edwards, A, Weigelt, J, Sundstrom, M, Doyle, D.A, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-05-30 | | Release date: | 2007-06-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Unusual binding interactions in PDZ domain crystal structures help explain binding mechanisms
Protein Sci., 19, 2010
|
|
4CYN
 
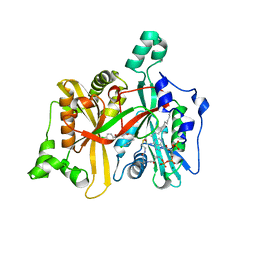 | | Leishmania major N-myristoyltransferase in complex with an aminoacylpyrrolidine inhibitor (2b) | | Descriptor: | (3R)-3-amino-4-(4-chlorophenyl)-1-[(3R,4S)-3-(4-chlorophenyl)-4-(hydroxymethyl)pyrrolidin-1-yl]butan-1-one, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, MAGNESIUM ION, ... | | Authors: | Hutton, J.A, Goncalves, V, Brannigan, J.A, Paape, D, Waugh, T, Roberts, S.M, Bell, A.S, Wilkinson, A.J, Smith, D.F, Leatherbarrow, R.J, Tate, E.W. | | Deposit date: | 2014-04-14 | | Release date: | 2014-10-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-Based Design of Potent and Selective Leishmania N- Myristoyltransferase Inhibitors.
J.Med.Chem., 57, 2014
|
|
2Q3W
 
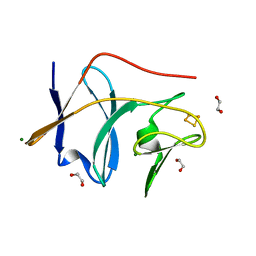 | | Ensemble refinement of the protein crystal structure of the cys84ala cys85ala double mutant of the [2Fe-2S] ferredoxin subunit of toluene-4-monooxygenase from Pseudomonas mendocina KR1 | | Descriptor: | 1,2-ETHANEDIOL, FE2/S2 (INORGANIC) CLUSTER, MAGNESIUM ION, ... | | Authors: | Levin, E.J, Kondrashov, D.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2007-05-30 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Ensemble refinement of protein crystal structures: validation and application.
Structure, 15, 2007
|
|
5D4A
 
 | | Crystal Structure of FABP4 in complex with 3-(2-phenyl-1H-indol-1-yl)propanoic acid | | Descriptor: | 3-(2-phenyl-1H-indol-1-yl)propanoic acid, Fatty acid-binding protein, adipocyte | | Authors: | Tagami, U, Takahashi, K, Igarashi, S, Ejima, C, Yoshida, T, Takeshita, S, Miyanaga, W, Sugiki, M, Tokumasu, M, Hatanaka, T, Kashiwagi, T, Ishikawa, K, Miyano, H, Mizukoshi, T. | | Deposit date: | 2015-08-07 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Interaction Analysis of FABP4 Inhibitors by X-ray Crystallography and Fragment Molecular Orbital Analysis
Acs Med.Chem.Lett., 7, 2016
|
|
4CPY
 
 | | Structure of the Neuraminidase from the B/Lyon/CHU/15.216/2011 virus in complex with Oseltamivir | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Vachieri, S.G, Collins, P.J, Escuret, V, Casalegno, J.S, Cattle, N, Ferraris, O, Sabatier, M, Frobert, E, Caro, V, Skehel, J.J, Gamblin, S.J, Valla, F, Valette, M, Ottmann, M, McCauley, J.W, Daniels, R.S, Lina, B. | | Deposit date: | 2014-02-09 | | Release date: | 2014-05-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Novel I221 L Substitution in Neuraminidase Confers High Level Resistance to Oseltamivir in Influenza B Viruses.
J.Infect.Dis., 210, 2014
|
|
5PP6
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 after initial refinement with no ligand modelled (structure 6) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 1, SODIUM ION | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PQA
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 after initial refinement with no ligand modelled (structure 47) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 1, SODIUM ION | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
2PJV
 
 | |
5PQQ
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 after initial refinement with no ligand modelled (structure 63) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 1, SODIUM ION | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
2PKH
 
 | | Structural Genomics, the crystal structure of the C-terminal domain of histidine utilization repressor from Pseudomonas syringae pv. tomato str. DC3000 | | Descriptor: | 1,2-ETHANEDIOL, Histidine utilization repressor | | Authors: | Tan, K, Zhou, M, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-04-17 | | Release date: | 2007-05-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The crystal structure of the C-terminal domain of histidine utilization repressor from Pseudomonas syringae pv. tomato str. DC3000.
To be Published
|
|
5PR6
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 after initial refinement with no ligand modelled (structure 79) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 1, SODIUM ION | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
4GGB
 
 | | Crystal structure of a proposed galactarolactone cycloisomerase from agrobacterium tumefaciens, TARGET EFI-500704, WITH BOUND CA, DISORDERED LOOPS | | Descriptor: | CALCIUM ION, CHLORIDE ION, Putative uncharacterized protein | | Authors: | Vetting, M.W, Bouvier, J.T, Morisco, L.L, Wasserman, S.R, Sojitra, S, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-08-06 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Purification, crystallization and structural elucidation of D-galactaro-1,4-lactone cycloisomerase from Agrobacterium tumefaciens involved in pectin degradation.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
5PRO
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 after initial refinement with no ligand modelled (structure 95) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 1, SODIUM ION | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
3AAD
 
 | | Structure of the histone chaperone CIA/ASF1-double bromodomain complex linking histone modifications and site-specific histone eviction | | Descriptor: | Histone chaperone ASF1A, SULFATE ION, Transcription initiation factor TFIID subunit 1 | | Authors: | Akai, Y, Adachi, N, Hayashi, Y, Eitoku, M, Sano, N, Natsume, R, Kudo, N, Tanokura, M, Senda, T, Horikoshi, M. | | Deposit date: | 2009-11-16 | | Release date: | 2010-04-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of the histone chaperone CIA/ASF1-double bromodomain complex linking histone modifications and site-specific histone eviction
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
5PS0
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 after initial refinement with no ligand modelled (structure 107) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 1, SODIUM ION | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PSH
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 after initial refinement with no ligand modelled (structure 125) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 1, SODIUM ION | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.43 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PSX
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 after initial refinement with no ligand modelled (structure 141) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 1, SODIUM ION | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PTS
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 after initial refinement with no ligand modelled (structure 170) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 1, SODIUM ION | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PU8
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 after initial refinement with no ligand modelled (structure 186) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 1, SODIUM ION | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
