2V93
 
 | | EQUILLIBRIUM MIXTURE OF OPEN AND PARTIALLY-CLOSED SPECIES IN THE APO STATE OF MALTODEXTRIN-BINDING PROTEIN BY PARAMAGNETIC RELAXATION ENHANCEMENT NMR | | Descriptor: | 1-(1-HYDROXY-2,2,6,6-TETRAMETHYLPIPERIDIN-4-YL)PYRROLIDINE-2,5-DIONE, MALTOSE-BINDING PERIPLASMIC PROTEIN | | Authors: | Clore, G.M, Tang, C. | | Deposit date: | 2007-08-21 | | Release date: | 2007-11-06 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Open-to-Closed Transition in Apo Maltose-Binding Protein Observed by Paramagnetic NMR.
Nature, 449, 2007
|
|
4RWH
 
 | | Crystal structure of T cell costimulatory ligand B7-1 (CD80) | | Descriptor: | T-lymphocyte activation antigen CD80 | | Authors: | Fedorov, A.A, Fedorov, E.V, Samanta, D, Hillerich, B, Seidel, R.D, Almo, S.C. | | Deposit date: | 2014-12-04 | | Release date: | 2014-12-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Crystal structure of T cell costimulatory ligand B7-1 (CD80)
To be Published
|
|
5SWR
 
 | | Crystal Structure of PI3Kalpha in complex with fragments 20 and 26 | | Descriptor: | 2-HYDROXYBENZOIC ACID, 6-hydroxy-3,4-dihydronaphthalen-1(2H)-one, CHLORIDE ION, ... | | Authors: | Gabelli, S.B, Vogelstein, B, Miller, M.S, Amzel, L.M. | | Deposit date: | 2016-08-08 | | Release date: | 2017-02-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Identification of allosteric binding sites for PI3K alpha oncogenic mutant specific inhibitor design.
Bioorg. Med. Chem., 25, 2017
|
|
4RXG
 
 | | Fructose-6-phosphate aldolase Q59E from E.coli | | Descriptor: | Fructose-6-phosphate aldolase 1, GLYCEROL, PENTAETHYLENE GLYCOL | | Authors: | Stellmacher, L, Sandalova, T, Leptihn, S, Schneider, G, Sprenger, G.A, Samland, A.K. | | Deposit date: | 2014-12-11 | | Release date: | 2015-10-07 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.154 Å) | | Cite: | Acid Base Catalyst Discriminates between a Fructose 6-Phosphate Aldolase and a Transaldolase
ChemCatChem, 2015
|
|
5SXI
 
 | | Crystal Structure of PI3Kalpha in complex with fragment 13 | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform, trans-cyclohexane-1,4-diol | | Authors: | Gabelli, S.B, Vogelstein, B, Miller, M.S, Amzel, L.M. | | Deposit date: | 2016-08-09 | | Release date: | 2017-02-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Identification of allosteric binding sites for PI3K alpha oncogenic mutant specific inhibitor design.
Bioorg. Med. Chem., 25, 2017
|
|
5WNM
 
 | |
3G2D
 
 | | Complex of Mth0212 and a 4 bp dsDNA with 3'-overhang | | Descriptor: | 5'-D(*CP*CP*TP*GP*UP*GP*CP*GP*AP*T)-3', 5'-D(*CP*GP*CP*G*CP*AP*GP*GP*C)-3', Exodeoxyribonuclease, ... | | Authors: | Lakomek, K, Dickmanns, A, Ficner, R. | | Deposit date: | 2009-01-31 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure Analysis of DNA Uridine Endonuclease Mth212 Bound to DNA
J.Mol.Biol., 399, 2010
|
|
4AIU
 
 | | A complex structure of BtGH84 | | Descriptor: | (3AR,5R,6S,7R,7AR)-2,5-BIS(HYDROXYMETHYL)-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D][1,3]OXAZOLE-6,7-DIOL, CALCIUM ION, O-GLCNACASE BT_4395 | | Authors: | He, Y, Davies, G.J. | | Deposit date: | 2012-02-13 | | Release date: | 2012-06-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Metabolism of Vertebrate Amino Sugars with N-Glycolyl Groups: Intracellular Beta-O-Linked N-Glycolylglucosamine (Glcngc), Udp-Glcngc, and the Biochemical and Structural Rationale for the Substrate Tolerance of Beta-O-Linked Beta-N-Acetylglucosaminidase.
J.Biol.Chem., 287, 2012
|
|
5TMW
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the OBHS derivative, 4-acetamidophenyl (1S,2R,4S)-5,6-bis(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonate | | Descriptor: | 4-(acetylamino)phenyl (1S,2R,4S)-5,6-bis(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonate, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Erumbi, R, Srinivasan, S, Bruno, N.E, Nowak, J, Izard, T, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-13 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.286 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
3FUJ
 
 | |
1FFR
 
 | | CRYSTAL STRUCTURE OF CHITINASE A MUTANT Y390F COMPLEXED WITH HEXA-N-ACETYLCHITOHEXAOSE (NAG)6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHITINASE A | | Authors: | Papanikolau, Y, Prag, G, Tavlas, G, Vorgias, C.E, Oppenheim, A.B, Petratos, K. | | Deposit date: | 2000-07-26 | | Release date: | 2001-09-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High resolution structural analyses of mutant chitinase A complexes with substrates provide new insight into the mechanism of catalysis.
Biochemistry, 40, 2001
|
|
5TM6
 
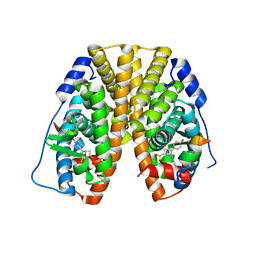 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the OBHS-ASC compound, 6-(4-((1R,4S,6R)-6-((4-bromophenoxy)sulfonyl)-3-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-2-en-2-yl)phenoxy)hexanoic acid | | Descriptor: | 6-{4-[(1S,4S,6R)-6-[(4-bromophenoxy)sulfonyl]-3-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-2-en-2-yl]phenoxy}hexanoic acid, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Erumbi, R, Srinivasan, S, Bruno, N.E, Nowak, J, Izard, T, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-12 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.542 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
3K0Z
 
 | |
5TCO
 
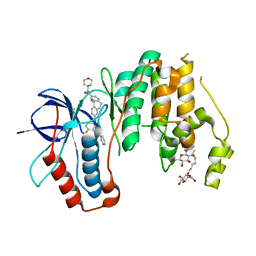 | | Human p38 MAP Kinase in Complex with Dibenzosuberone Compound 1 | | Descriptor: | 3-[(3-benzamido-4-fluoranyl-phenyl)amino]-~{N}-(2-morpholin-4-ylethyl)-11-oxidanylidene-5,6-dihydrodibenzo[1,2-~{d}:1',2'-~{f}][7]annulene-9-carboxamide, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Mayer-Wrangowski, S.C, Rauh, D. | | Deposit date: | 2016-09-15 | | Release date: | 2017-04-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Optimized Target Residence Time: Type I1/2 Inhibitors for p38 alpha MAP Kinase with Improved Binding Kinetics through Direct Interaction with the R-Spine.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5QCP
 
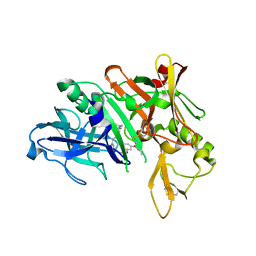 | | Crystal structure of BACE complex with BMC018 | | Descriptor: | (4S)-4-[(1R)-1-hydroxy-2-({[3-(propan-2-yl)phenyl]methyl}amino)ethyl]-19-(2-oxopropoxy)-11,16-dioxa-3-azatricyclo[15.3.1.1~6,10~]docosa-1(21),6(22),7,9,17,19-hexaen-2-one, Beta-secretase 1 | | Authors: | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-12-01 | | Release date: | 2020-06-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
5QCX
 
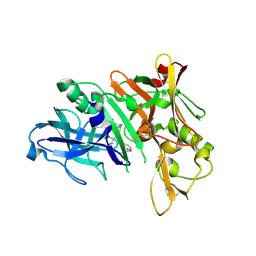 | | Crystal structure of BACE complex with BMC007 | | Descriptor: | (9R,11S)-3-ethyl-11-[(1R)-1-hydroxy-2-({[3-(propan-2-yl)phenyl]methyl}amino)ethyl]-9-methyl-3,12-diazabicyclo[12.3.1]octadeca-1(18),14,16-triene-2,13-dione, Beta-secretase 1 | | Authors: | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-12-01 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
5QD9
 
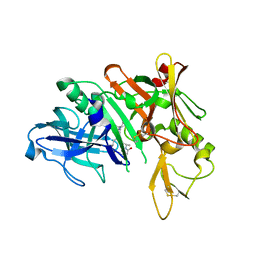 | | Crystal structure of BACE complex with BMC005 | | Descriptor: | (5S,8S,10R)-8-[(1R)-2-{[1-(3-tert-butylphenyl)cyclopropyl]amino}-1-hydroxyethyl]-4,5,10-trimethyl-1-oxa-4,7-diazacyclohexadecane-3,6-dione, Beta-secretase 1 | | Authors: | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-12-01 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
5QDP
 
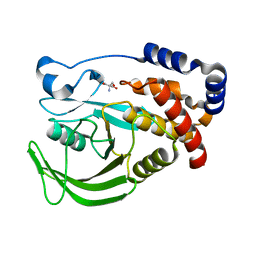 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with compound_FMOPL000207a | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, N-ethyl-N'-(5-methyl-1,2-oxazol-3-yl)urea, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2018-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.744 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
5QD5
 
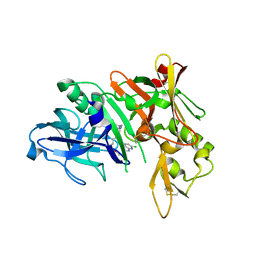 | | Crystal structure of BACE complex with BMC009 | | Descriptor: | (10S,12S)-17-chloro-12-[(1R)-1-hydroxy-2-({[3-(propan-2-yl)phenyl]methyl}amino)ethyl]-10-methyl-7-oxa-2,13,18-triazabicyclo[13.3.1]nonadeca-1(19),15,17-trien-14-one, Beta-secretase 1 | | Authors: | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-12-01 | | Release date: | 2020-06-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
5QDT
 
 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with compound_FMOPL000475a | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1, ~{N}-(1-propyl-1,2,3,4-tetrazol-5-yl)furan-2-carboxamide | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2018-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.821 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
5QEB
 
 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with compound_FMOPL000639a | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-chloro-N~1~-(pyridin-4-yl)benzene-1,2-diamine, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2018-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.733 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
5QEL
 
 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with compound_FMSOA000675b | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-(pyrrolidin-1-yl)pyridine-2-carbonitrile, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2018-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.654 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
6R9K
 
 | |
4JC1
 
 | |
5QFH
 
 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with compound_FMOOA000525a | | Descriptor: | (5S,8R,8aS)-8-hydroxyhexahydro-3H-5,8-ethano[1,3]oxazolo[3,4-a]pyridin-3-one, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2018-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.591 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
