5D7Y
 
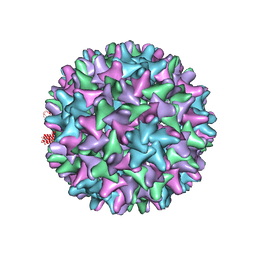 | | Crystal structure of Hepatitis B virus T=4 capsid in complex with the allosteric modulator HAP18 | | Descriptor: | Capsid protein, methyl (4R)-4-(2-chloro-4-fluorophenyl)-6-{[4-(3-hydroxypenta-1,4-diyn-3-yl)piperidin-1-yl]methyl}-2-(pyridin-2-yl)-1,4-dihydropyrimidine-5-carboxylate | | Authors: | Venkatakrishnan, B, Katen, S.P, Zlotnick, A. | | Deposit date: | 2015-08-14 | | Release date: | 2016-02-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.894 Å) | | Cite: | Hepatitis B Virus Capsids Have Diverse Structural Responses to Small-Molecule Ligands Bound to the Heteroaryldihydropyrimidine Pocket.
J.Virol., 90, 2016
|
|
1CR7
 
 | | PEANUT LECTIN-LACTOSE COMPLEX MONOCLINIC FORM | | Descriptor: | CALCIUM ION, LECTIN, MANGANESE (II) ION, ... | | Authors: | Ravishankar, R, Suguna, K, Surolia, A, Vijayan, M. | | Deposit date: | 1999-08-14 | | Release date: | 2001-04-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of the peanut lectin-lactose complex at acidic pH: retention of unusual quaternary structure, empty and carbohydrate bound combining sites, molecular mimicry and crystal packing directed by interactions at the combining site.
Proteins, 43, 2001
|
|
5D38
 
 | |
6Y2S
 
 | |
4PL4
 
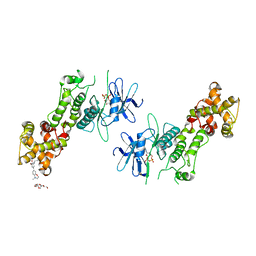 | | Crystal structure of murine IRE1 in complex with OICR464 inhibitor | | Descriptor: | 2,5,8,11,14,17,20,23,26,29,32,35,38,41,44,47,50,53,56,59,62,65,68,71,74,77,80-HEPTACOSAOXADOOCTACONTAN-82-OL, 2-methoxy-6-methyl-4-(4-methyl-3,4-dihydro-2H-1,4-benzoxazin-7-yl)phenol, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Sanches, M, Duffy, N, Talukdar, M, Thevakumaran, N, Chiovitti, D, Al-awar, R, Patterson, J.B, Sicheri, F. | | Deposit date: | 2014-05-16 | | Release date: | 2014-09-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and mechanism of action of the hydroxy-aryl-aldehyde class of IRE1 endoribonuclease inhibitors.
Nat Commun, 5, 2014
|
|
3QHT
 
 | | Crystal Structure of the Monobody ySMB-1 bound to yeast SUMO | | Descriptor: | GLYCEROL, Monobody ySMB-1, Ubiquitin-like protein SMT3 | | Authors: | Koide, S, Gilbreth, R.N. | | Deposit date: | 2011-01-26 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Isoform-specific monobody inhibitors of small ubiquitin-related modifiers engineered using structure-guided library design.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4KA7
 
 | | Structure of Organellar OligoPeptidase (E572Q) in complex with an endogenous substrate | | Descriptor: | CHLORIDE ION, GLYCEROL, Oligopeptidase A, ... | | Authors: | Berntsson, R.P.-A, Kmiec, B, Teixeira, P.F, Svensson, L.M, Bakali, A, Glaser, E, Stenmark, P. | | Deposit date: | 2013-04-22 | | Release date: | 2013-09-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Organellar oligopeptidase (OOP) provides a complementary pathway for targeting peptide degradation in mitochondria and chloroplasts.
Proc. Natl. Acad. Sci. U.S.A., 110, 2013
|
|
4KC7
 
 | | Crystal Structure of Endo-1,5-alpha-L-arabinanase from Thermotoga petrophila RKU-1 | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, Glycoside hydrolase, ... | | Authors: | Nascimento, A.F.Z, Polo, C.C, Santos, C.R, Costa, M.C.M.F, Mesa, A.N, Prade, R.A, Ruller, R, Squina, F.M, Murakami, M.T. | | Deposit date: | 2013-04-24 | | Release date: | 2014-02-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Mechanistic strategies for catalysis adopted by evolutionary distinct family 43 arabinanases.
J.Biol.Chem., 289, 2014
|
|
2FEK
 
 | | Structure of a protein tyrosine phosphatase | | Descriptor: | Low molecular weight protein-tyrosine-phosphatase wzb | | Authors: | Lescop, E, Jin, C. | | Deposit date: | 2005-12-16 | | Release date: | 2006-05-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The solution structure of Escherichia coli Wzb reveals a novel substrate recognition mechanism of prokaryotic low molecular weight protein-tyrosine phosphatases
J.Biol.Chem., 281, 2006
|
|
2A0U
 
 | |
4FN4
 
 | | short-chain NAD(H)-dependent dehydrogenase/reductase from Sulfolobus acidocaldarius | | Descriptor: | GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Pennacchio, A, Sannino, V, Sorrentino, G, Rossi, M, Raia, C.A, Esposito, L. | | Deposit date: | 2012-06-19 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Biochemical and structural characterization of recombinant short-chain NAD(H)-dependent dehydrogenase/reductase from Sulfolobus acidocaldarius highly enantioselective on diaryl diketone benzil.
Appl.Microbiol.Biotechnol., 97, 2013
|
|
5A8Q
 
 | | Crystal structure beta-glucanase SdGluc5_26A from Saccharophagus degradans in complex with tetrasaccharide A obtained by soaking | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Sulzenbacher, G, Lafond, M, Freyd, T, Henrissat, B, Coutinho, R.M, Berrin, J.G, Garron, M.L. | | Deposit date: | 2015-07-16 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Quaternary Structure of a Glycoside Hydrolase Dictates Specificity Towards Beta-Glucans
J.Biol.Chem., 291, 2016
|
|
6QXU
 
 | | Human TNKS1 in complex with 6,8-Difluoro-2-[4-(1-hydroxy-1-methyl-ethyl)-phenyl]-3H-quinazolin-4-one | | Descriptor: | 6,8-bis(fluoranyl)-2-[4-(2-oxidanylpropan-2-yl)phenyl]-3~{H}-quinazolin-4-one, BETA-MERCAPTOETHANOL, Poly [ADP-ribose] polymerase tankyrase-1, ... | | Authors: | Musil, D, Lehmann, D, Buchstaller, H.-P. | | Deposit date: | 2019-03-08 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Discovery and Optimization of 2-Arylquinazolin-4-ones into a Potent and Selective Tankyrase Inhibitor Modulating Wnt Pathway Activity.
J.Med.Chem., 62, 2019
|
|
5A8P
 
 | | Crystal structure beta-glucanase SdGluc5_26A from Saccharophagus degradans in complex with tetrasaccharide B | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Sulzenbacher, G, Lafond, M, Freyd, T, Henrissat, B, Coutinho, R.M, Berrin, J.G, Garron, M.L. | | Deposit date: | 2015-07-16 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Quaternary Structure of a Glycoside Hydrolase Dictates Specificity Towards Beta-Glucans
J.Biol.Chem., 291, 2016
|
|
5AMB
 
 | | Crystal structure of the Angiotensin-1 converting enzyme N-domain in complex with amyloid-beta 35-42 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, AMYLOID BETA A4 PROTEIN, ANGIOTENSIN-CONVERTING ENZYME, ... | | Authors: | Masuyer, G, Larmuth, K.M, Douglas, R.G, Sturrock, E.D, Acharya, K.R. | | Deposit date: | 2015-03-10 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The Kinetic and Structural Characterisation of Amyloid-Beta Metabolism by Human Angiotensin-1- Converting Enzyme (Ace)
FEBS J., 283, 2016
|
|
5ARL
 
 | |
5AMC
 
 | | Crystal structure of the Angiotensin-1 converting enzyme N-domain in complex with amyloid-beta fluorogenic fragment 4-10 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ANGIOTENSIN-CONVERTING ENZYME, CHLORIDE ION, ... | | Authors: | Masuyer, G, Larmuth, K.M, Douglas, R.G, Sturrock, E.D, Acharya, K.R. | | Deposit date: | 2015-03-10 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Kinetic and Structural Characterisation of Amyloid-Beta Metabolism by Human Angiotensin-1- Converting Enzyme (Ace)
FEBS J., 283, 2016
|
|
3GAL
 
 | |
3RP7
 
 | | Crystal Structure of Klebsiella pneumoniae HpxO complexed with FAD and uric acid | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, URIC ACID, flavoprotein monooxygenase | | Authors: | Hicks, K.A, O'Leary, S.E, Begley, T.P, Ealick, S.E. | | Deposit date: | 2011-04-26 | | Release date: | 2012-05-16 | | Last modified: | 2013-02-06 | | Method: | X-RAY DIFFRACTION (2.042 Å) | | Cite: | Structural and Mechanistic Studies of HpxO, a Novel Flavin Adenine Dinucleotide-Dependent Urate Oxidase from Klebsiella pneumoniae.
Biochemistry, 52, 2013
|
|
5BNF
 
 | | Apo structure of porcine CD38 | | Descriptor: | Uncharacterized protein | | Authors: | Ting, K.Y, Leung, C.P.F, Graeff, R.M, Lee, H.C, Hao, Q, Kotaka, M. | | Deposit date: | 2015-05-26 | | Release date: | 2016-05-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Porcine CD38 exhibits prominent secondary NAD(+) cyclase activity.
Protein Sci., 25, 2016
|
|
4E4K
 
 | | Crystal Structure of PPARgamma with the ligand JO21 | | Descriptor: | (2S)-3-phenyl-2-{[2'-(propan-2-yl)biphenyl-4-yl]oxy}propanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Pochetti, G, Montanari, R, Loiodice, F, Fracchiolla, G, Laghezza, A, Carbonara, G, Piemontese, L, Lavecchia, A, Novellino, E. | | Deposit date: | 2012-03-13 | | Release date: | 2013-01-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | New 2-(Aryloxy)-3-phenylpropanoic Acids as Peroxisome Proliferator-Activated Receptor alpha/gamma Dual Agonists Able To Upregulate Mitochondrial Carnitine Shuttle System Gene Expression.
J.Med.Chem., 56, 2013
|
|
4E4Q
 
 | | Crystal structure of PPARgamma with the ligand FS214 | | Descriptor: | (2R)-3-phenyl-2-{[2'-(propan-2-yl)biphenyl-4-yl]oxy}propanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Pochetti, G, Montanari, R, Loiodice, F, Fracchiolla, G, Laghezza, A, Carbonara, G, Piemontese, L, Lavecchia, A, Novellino, E. | | Deposit date: | 2012-03-13 | | Release date: | 2013-01-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | New 2-(Aryloxy)-3-phenylpropanoic Acids as Peroxisome Proliferator-Activated Receptor alpha/gamma Dual Agonists Able To Upregulate Mitochondrial Carnitine Shuttle System Gene Expression.
J.Med.Chem., 56, 2013
|
|
4K7U
 
 | | Crystal structure of Zn2.3-hUb (human ubiquitin) adduct from a solution 70 mM zinc acetate/1.3 mM hUb | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Fermani, S, Falini, G, Calvaresi, M, Bottoni, A, Arnesano, F, Natile, G. | | Deposit date: | 2013-04-17 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Conformational selection of ubiquitin quaternary structures driven by zinc ions.
Chemistry, 19, 2013
|
|
4K6O
 
 | | X-ray structure uridine phosphorylase from Vibrio cholerae in complex with 6-methyluracil at 1.17 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-methylpyrimidine-2,4-diol, ... | | Authors: | Prokofev, I.I, Lashkov, A.A, Gabdoulkhakov, A.G, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2013-04-16 | | Release date: | 2014-05-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Crystallization and preliminary X-ray study of Vibrio cholerae uridine phosphorylase in complex with 6-methyluracil.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
5ARJ
 
 | |
