6D3P
 
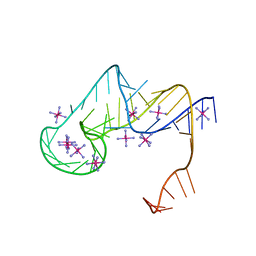 | | Crystal structure of an exoribonuclease-resistant RNA from Sweet clover necrotic mosaic virus (SCNMV) | | Descriptor: | IRIDIUM HEXAMMINE ION, RNA (45-MER) | | Authors: | Steckelberg, A.-L, Akiyama, B.M, Costantino, D.A, Sit, T.L, Nix, J.C, Kieft, J.S. | | Deposit date: | 2018-04-16 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A folded viral noncoding RNA blocks host cell exoribonucleases through a conformationally dynamic RNA structure.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2BTV
 
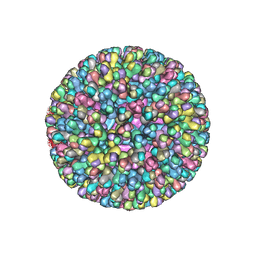 | | ATOMIC MODEL FOR BLUETONGUE VIRUS (BTV) CORE | | Descriptor: | PROTEIN (VP3 CORE PROTEIN), PROTEIN (VP7 CORE PROTEIN) | | Authors: | Grimes, J.M, Burroughs, J.N, Gouet, P, Diprose, J.M, Malby, R, Zientras, S, Mertens, P.P.C, Stuart, D.I. | | Deposit date: | 1998-09-05 | | Release date: | 1998-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The atomic structure of the bluetongue virus core.
Nature, 395, 1998
|
|
6D43
 
 | |
6CEN
 
 | | Crystal Structure of WHSC1L1 in Complex with Inhibitor PEP21 | | Descriptor: | ACE-GLY-VAL-NLE-ARG-ILE-NH2, Histone-lysine N-methyltransferase NSD3, S-ADENOSYLMETHIONINE, ... | | Authors: | Boriack-Sjodin, P.A, Swinger, K, Farrow, N.A. | | Deposit date: | 2018-02-12 | | Release date: | 2018-05-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Identification of a peptide inhibitor for the histone methyltransferase WHSC1.
PLoS ONE, 13, 2018
|
|
2C1A
 
 | | Structure of cAMP-dependent protein kinase complexed with Isoquinoline-5-sulfonic acid (2-(2-(4-chlorobenzyloxy)ethylamino) ethyl)amide | | Descriptor: | CAMP-DEPENDENT PROTEIN KINASE, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR, ISOQUINOLINE-5-SULFONIC ACID (2-(2-(4-CHLOROBENZYLOXY)ETHYLAMINO)ETHYL)AMIDE | | Authors: | Collins, I, Caldwell, J, Fonseca, T, Donald, A, Bavetsias, V, Hunter, L.J, Garrett, M.D, Rowlands, M.G, Aherne, G.W, Davies, T.G, Berdini, V, Woodhead, S.J, Seavers, L.C.A, Wyatt, P.G, Workman, P, McDonald, E. | | Deposit date: | 2005-09-12 | | Release date: | 2005-11-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-based design of isoquinoline-5-sulfonamide inhibitors of protein kinase B.
Bioorg. Med. Chem., 14, 2006
|
|
2C1B
 
 | | Structure of cAMP-dependent protein kinase complexed with (4R,2S)-5'-(4-(4-Chlorobenzyloxy)pyrrolidin-2-ylmethanesulfonyl)isoquinoline | | Descriptor: | (4R,2S)-5'-(4-(4-CHLOROBENZYLOXY)PYRROLIDIN-2-YLMETHANESULFONYL)ISOQUINOLINE, CAMP-DEPENDENT PROTEIN KINASE, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR | | Authors: | Collins, I, Caldwell, J, Fonseca, T, Donald, A, Bavetsias, V, Hunter, L.J, Garrett, M.D, Rowlands, M.G, Aherne, G.W, Davies, T.G, Berdini, V, Woodhead, S.J, Seavers, L.C.A, Wyatt, P.G, Workman, P, McDonald, E. | | Deposit date: | 2005-09-12 | | Release date: | 2005-11-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design of isoquinoline-5-sulfonamide inhibitors of protein kinase B.
Bioorg. Med. Chem., 14, 2006
|
|
2C3C
 
 | | 2.01 Angstrom X-ray crystal structure of a mixed disulfide between coenzyme M and NADPH-dependent oxidoreductase 2-ketopropyl coenzyme M carboxylase | | Descriptor: | 1-THIOETHANESULFONIC ACID, 2-OXOPROPYL-COM REDUCTASE, ACETONE, ... | | Authors: | Pandey, A.S, Nocek, B, Clark, D.D, Ensign, S.A, Peters, J.W. | | Deposit date: | 2005-10-05 | | Release date: | 2005-12-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Mechanistic Implications of the Structure of the Mixed-Disulfide Intermediate of the Disulfide Oxidoreductase, 2-Ketopropyl-Coenzyme M Oxidoreductase/Carboxylase.
Biochemistry, 45, 2006
|
|
6CFD
 
 | | ADEP4 bound to E. faecium ClpP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ATP-dependent Clp protease proteolytic subunit, N-[(6aS,12S,15aS,17R,21R,23aS)-17,21-dimethyl-6,11,15,20,23-pentaoxooctadecahydro-2H,6H,11H,15H-pyrido[2,1-i]dipyrrolo[2,1-c:2',1'-l][1,4,7,10,13]oxatetraazacyclohexadecin-12-yl]-3,5-difluoro-Nalpha-[(2E)-hept-2-enoyl]-L-phenylalaninamide | | Authors: | Lee, R.E, Griffith, E.C. | | Deposit date: | 2018-02-14 | | Release date: | 2018-05-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | In VivoandIn VitroEffects of a ClpP-Activating Antibiotic against Vancomycin-Resistant Enterococci.
Antimicrob. Agents Chemother., 62, 2018
|
|
6CWO
 
 | |
2BFC
 
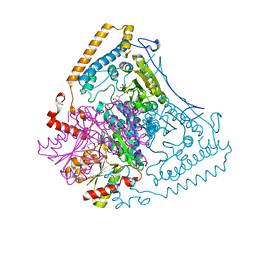 | | Reactivity modulation of human branched-chain alpha-ketoacid dehydrogenase by an internal molecular switch | | Descriptor: | 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, 2-{3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-4-METHYL-2-OXO-2,3-DIHYDRO-1,3-THIAZOL-5-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, ... | | Authors: | Machius, M, Wynn, R.M, Chuang, J.L, Tomchick, D.R, Brautigam, C.A, Chuang, D.T. | | Deposit date: | 2004-12-06 | | Release date: | 2006-02-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | A Versatile Conformational Switch Regulates Reactivity in Human Branched-Chain Alpha-Ketoacid Dehydrogenase.
Structure, 14, 2006
|
|
2BIO
 
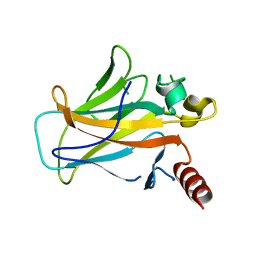 | |
6CWZ
 
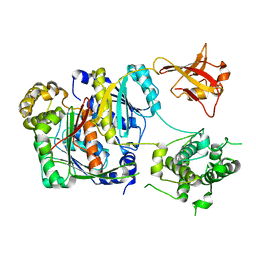 | | Crystal structure of apo SUMO E1 | | Descriptor: | SUMO-activating enzyme subunit 1, SUMO-activating enzyme subunit 2, ZINC ION | | Authors: | Lv, Z, Yuan, L, Atkison, J.H, Williams, K.M, Olsen, S.K. | | Deposit date: | 2018-04-01 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Molecular mechanism of a covalent allosteric inhibitor of SUMO E1 activating enzyme.
Nat Commun, 9, 2018
|
|
2C5S
 
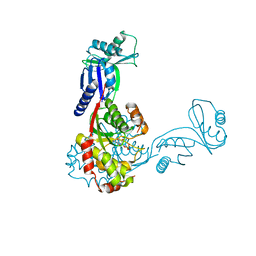 | | Crystal structure of Bacillus anthracis ThiI, a tRNA-modifying enzyme containing the predicted RNA-binding THUMP domain | | Descriptor: | ADENOSINE MONOPHOSPHATE, PROBABLE THIAMINE BIOSYNTHESIS PROTEIN THII | | Authors: | Waterman, D.G, Ortiz-Lombardia, M, Fogg, M.J, Koonin, E.V, Antson, A.A. | | Deposit date: | 2005-11-01 | | Release date: | 2005-11-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Bacillus anthracis ThiI, a tRNA-modifying enzyme containing the predicted RNA-binding THUMP domain.
J.Mol.Biol., 356, 2006
|
|
2C3M
 
 | | Crystal Structure Of Pyruvate-Ferredoxin Oxidoreductase From Desulfovibrio africanus | | Descriptor: | CALCIUM ION, CHLORIDE ION, IRON/SULFUR CLUSTER, ... | | Authors: | Cavazza, C, Contreras-Martel, C, Pieulle, L, Chabriere, E, Hatchikian, E.C, Fontecilla-Camps, J.C. | | Deposit date: | 2005-10-11 | | Release date: | 2006-02-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Flexibility of Thiamine Diphosphate Revealed by Kinetic Crystallographic Studies of the Reaction of Pyruvate-Ferredoxin Oxidoreductase with Pyruvate.
Structure, 14, 2006
|
|
2C6E
 
 | | Aurora A kinase activated mutant (T287D) in complex with a 5- aminopyrimidinyl quinazoline inhibitor | | Descriptor: | N-{5-[(7-{[(2S)-2-HYDROXY-3-PIPERIDIN-1-YLPROPYL]OXY}-6-METHOXYQUINAZOLIN-4-YL)AMINO]PYRIMIDIN-2-YL}BENZAMIDE, SERINE/THREONINE-PROTEIN KINASE 6 | | Authors: | Pauptit, R.A, Pannifer, A.D, Breed, J, McMiken, H.H.J, Rowsell, S, Anderson, M. | | Deposit date: | 2005-11-09 | | Release date: | 2006-01-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Sar and Inhibitor Complex Structure Determination of a Novel Class of Potent and Specific Aurora Kinase Inhibitors.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
6CY3
 
 | | Horse liver E267N alcohol dehydrogenase complex with 3'-dephosphocoenzyme A | | Descriptor: | DEPHOSPHO COENZYME A, ZINC ION, alcohol dehydrogenase | | Authors: | Plapp, B.V. | | Deposit date: | 2018-04-04 | | Release date: | 2018-04-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Substitutions of a buried glutamate residue hinder the conformational change in horse liver alcohol dehydrogenase and yield a surprising complex with endogenous 3'-Dephosphocoenzyme A.
Arch. Biochem. Biophys., 653, 2018
|
|
6CI8
 
 | |
2BFI
 
 | | Molecular basis for amyloid fibril formation and stability | | Descriptor: | SYNTHETIC PEPTIDE | | Authors: | Makin, O.S, Atkins, E, Sikorski, P, Johansson, J, Serpell, L.C. | | Deposit date: | 2004-12-07 | | Release date: | 2005-01-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Molecular Basis for Amyloid Fibril Formation and Stability
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2C2H
 
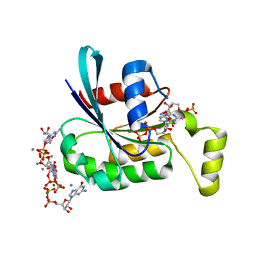 | | CRYSTAL STRUCTURE OF THE HUMAN RAC3 IN COMPLEX WITH GDP | | Descriptor: | CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Debreczeni, J.E, Yang, X, Zao, Y, Elkins, J, Gileadi, C, Burgess, N, Colebrook, S, Gileadi, O, Fedorov, O, Bunkoczi, G, von Delft, F, Doyle, D, Sundstrom, M, Arrowsmith, C, Weigelt, J, Edwards, A. | | Deposit date: | 2005-09-29 | | Release date: | 2005-10-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of the Human Rac3 in Complex with Gdp
To be Published
|
|
2C0F
 
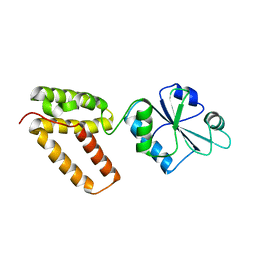 | | Structure of Wind Y53F mutant | | Descriptor: | WINDBEUTEL PROTEIN | | Authors: | Sevvana, M, Ma, Q, Barnewitz, K, Guo, C, Soling, H.-D, Ferrari, D.M, Sheldrick, G.M. | | Deposit date: | 2005-09-02 | | Release date: | 2006-08-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural Elucidation of the Pdi-Related Chaperone Wind with the Help of Mutants.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
6CYT
 
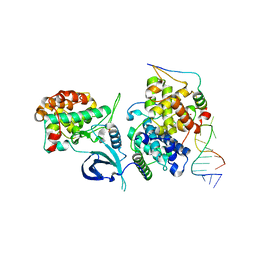 | | HIV-1 TAR loop in complex with Tat:AFF4:P-TEFb | | Descriptor: | AF4/FMR2 family member 4, Cyclin-T1, Cyclin-dependent kinase 9, ... | | Authors: | Schulze Gahmen, U, Hurley, J.H. | | Deposit date: | 2018-04-06 | | Release date: | 2018-12-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural mechanism for HIV-1 TAR loop recognition by Tat and the super elongation complex.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2BGI
 
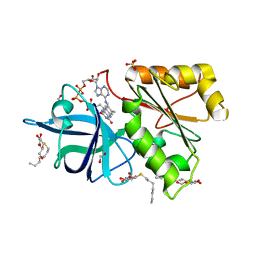 | | X-Ray Structure of the Ferredoxin-NADP(H) Reductase from Rhodobacter capsulatus complexed with three molecules of the detergent n-heptyl- beta-D-thioglucoside at 1.7 Angstroms | | Descriptor: | CARBON DIOXIDE, FERREDOXIN-NADP(H) REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Perez-Dorado, J.I, Hermoso, J.A, Nogues, I, Frago, S, Bittel, C, Mayhew, S.G, Gomez-Moreno, C, Medina, M, Cortez, N, Carrillo, N. | | Deposit date: | 2004-12-23 | | Release date: | 2005-09-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | The Ferredoxin-Nadp(H) Reductase from Rhodobacter Capsulatus: Molecular Structure and Catalytic Mechanism
Biochemistry, 44, 2005
|
|
2BGU
 
 | | CRYSTAL STRUCTURE OF THE DNA MODIFYING ENZYME BETA-GLUCOSYLTRANSFERASE IN THE PRESENCE AND ABSENCE OF THE SUBSTRATE URIDINE DIPHOSPHOGLUCOSE | | Descriptor: | BETA-GLUCOSYLTRANSFERASE, URIDINE-5'-DIPHOSPHATE | | Authors: | Vrielink, A, Rueger, W, Driessen, H.P.C, Freemont, P.S. | | Deposit date: | 1994-06-09 | | Release date: | 1995-12-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the DNA modifying enzyme beta-glucosyltransferase in the presence and absence of the substrate uridine diphosphoglucose.
EMBO J., 13, 1994
|
|
2C4V
 
 | | H. pylori type II DHQase in complex with citrate | | Descriptor: | 3-DEHYDROQUINATE DEHYDRATASE, CITRIC ACID | | Authors: | Robinson, D.A, Lapthorn, A.J. | | Deposit date: | 2005-10-24 | | Release date: | 2006-02-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structures of Helicobacter Pylori Type II Dehydroquinase Inhibitor Complexes: New Directions for Inhibitor Design.
J.Med.Chem., 49, 2006
|
|
2C21
 
 | | Specificity of the Trypanothione-dependednt Leishmania major Glyoxalase I: Structure and biochemical comparison with the human enzyme | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, NICKEL (II) ION, ... | | Authors: | Ariza, A, Vickers, T.J, Greig, N, Armour, K.A, Eggleston, I.M, Fairlamb, A.H, Bond, C.S. | | Deposit date: | 2005-09-23 | | Release date: | 2006-02-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Specificity of the Trypanothione-Dependent Leishmania Major Glyoxalase I: Structure and Biochemical Comparison with the Human Enzyme.
Mol.Microbiol., 59, 2006
|
|
