5XD1
 
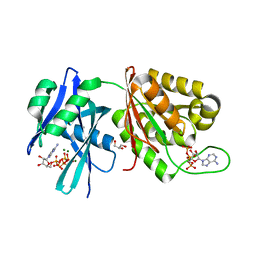 | | Crystal structure of Mycobacterium smegmatis MutT1 in complex with Ap5A, ATP and magnesium | | Descriptor: | ADENOSINE-5'-PENTAPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, ... | | Authors: | Arif, S.M, Varshney, U, Vijayan, M. | | Deposit date: | 2017-03-24 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Hydrolysis of diadenosine polyphosphates. Exploration of an additional role of Mycobacterium smegmatis MutT1
J. Struct. Biol., 199, 2017
|
|
5XD3
 
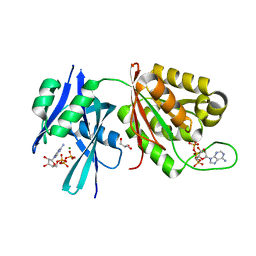 | | Crystal structure of Mycobacterium smegmatis MutT1 in complex with ATP (I) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Arif, S.M, Varshney, U, Vijayan, M. | | Deposit date: | 2017-03-24 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Hydrolysis of diadenosine polyphosphates. Exploration of an additional role of Mycobacterium smegmatis MutT1
J. Struct. Biol., 199, 2017
|
|
5YGU
 
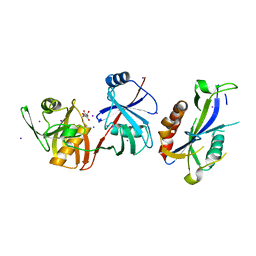 | | Crystal structure of Escherichia coli (strain K12) mRNA Decapping Complex RppH-DapF | | Descriptor: | Diaminopimelate epimerase, IODIDE ION, L(+)-TARTARIC ACID, ... | | Authors: | Wang, Q, Guan, Z.Y, Zhang, D.L, Zou, T.T, Yin, P. | | Deposit date: | 2017-09-27 | | Release date: | 2018-06-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.298 Å) | | Cite: | DapF stabilizes the substrate-favoring conformation of RppH to stimulate its RNA-pyrophosphohydrolase activity in Escherichia coli.
Nucleic Acids Res., 46, 2018
|
|
5XD2
 
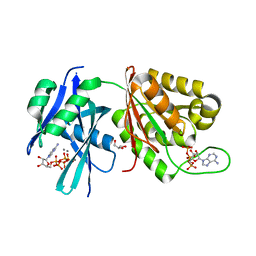 | | Crystal structure of Mycobacterium smegmatis MutT1 in complex with Ap5A, ATP and manganese | | Descriptor: | ADENOSINE-5'-PENTAPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, ... | | Authors: | Arif, S.M, Varshney, U, Vijayan, M. | | Deposit date: | 2017-03-24 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Hydrolysis of diadenosine polyphosphates. Exploration of an additional role of Mycobacterium smegmatis MutT1
J. Struct. Biol., 199, 2017
|
|
5XD5
 
 | | Crystal structure of Mycobacterium smegmatis MutT1 in complex with ATP, magnesium fluoride and phosphate | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, FLUORIDE ION, ... | | Authors: | Arif, S.M, Varshney, U, Vijayan, M. | | Deposit date: | 2017-03-24 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Hydrolysis of diadenosine polyphosphates. Exploration of an additional role of Mycobacterium smegmatis MutT1
J. Struct. Biol., 199, 2017
|
|
5XD4
 
 | | Crystal structure of Mycobacterium smegmatis MutT1 in complex with ATP (II) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Arif, S.M, Varshney, U, Vijayan, M. | | Deposit date: | 2017-03-24 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Hydrolysis of diadenosine polyphosphates. Exploration of an additional role of Mycobacterium smegmatis MutT1
J. Struct. Biol., 199, 2017
|
|
3CNG
 
 | | Crystal structure of NUDIX hydrolase from Nitrosomonas europaea | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Osipiuk, J, Xu, X, Zheng, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-03-25 | | Release date: | 2008-04-08 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray crystal structure of NUDIX hydrolase from Nitrosomonas europaea.
To be Published
|
|
4HFQ
 
 | | Crystal structure of UDP-X diphosphatase | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Duong-Ly, K.C, Amzel, L.M, Gabelli, S.B. | | Deposit date: | 2012-10-05 | | Release date: | 2013-08-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | A UDP-X diphosphatase from Streptococcus pneumoniae hydrolyzes precursors of peptidoglycan biosynthesis.
Plos One, 8, 2013
|
|
2A6T
 
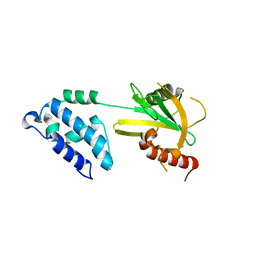 | |
2QJO
 
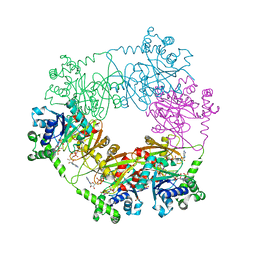 | | crystal structure of a bifunctional NMN adenylyltransferase/ADP ribose pyrophosphatase (NadM) complexed with ADPRP and NAD from Synechocystis sp. | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Bifunctional NMN adenylyltransferase/Nudix hydrolase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Huang, N, Sorci, L, Zhang, X, Brautigan, C, Raffaelli, N, Magni, G, Grishin, N.V, Osterman, A, Zhang, H. | | Deposit date: | 2007-07-08 | | Release date: | 2008-03-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Bifunctional NMN Adenylyltransferase/ADP-Ribose Pyrophosphatase: Structure and Function in Bacterial NAD Metabolism.
Structure, 16, 2008
|
|
2QJT
 
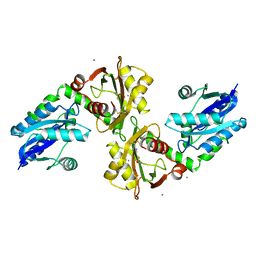 | | Crystal structure of a bifunctional NMN adenylyltransferase/ADP ribose pyrophosphatase complexed with AMP and MN ion from Francisella tularensis | | Descriptor: | ADENOSINE MONOPHOSPHATE, MANGANESE (II) ION, Nicotinamide-nucleotide adenylyltransferase | | Authors: | Huang, N, Sorci, L, Zhang, X, Brautigan, C, Raffaelli, N, Magni, G, Grishin, N.V, Osterman, A, Zhang, H. | | Deposit date: | 2007-07-09 | | Release date: | 2008-03-04 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Bifunctional NMN Adenylyltransferase/ADP-Ribose Pyrophosphatase: Structure and Function in Bacterial NAD Metabolism.
Structure, 16, 2008
|
|
2QKM
 
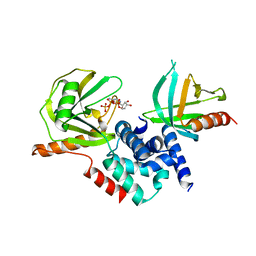 | |
2R5W
 
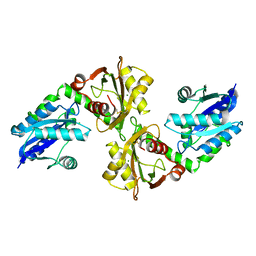 | | Crystal structure of a bifunctional NMN adenylyltransferase/ADP ribose pyrophosphatase from Francisella tularensis | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Nicotinamide-nucleotide adenylyltransferase | | Authors: | Huang, N, Sorci, L, Zhang, X, Brautigan, C, Li, X, Raffaelli, N, Grishin, N, Osterman, A, Zhang, H. | | Deposit date: | 2007-09-04 | | Release date: | 2008-03-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Bifunctional NMN Adenylyltransferase/ADP-Ribose Pyrophosphatase: Structure and Function in Bacterial NAD Metabolism.
Structure, 16, 2008
|
|
2FML
 
 | |
2GB5
 
 | |
5KQ1
 
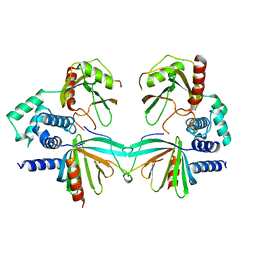 | | Crystal structure of S. pombe Dcp1/Dcp2 in complex with H. sapiens PNRC2 | | Descriptor: | Proline-rich nuclear receptor coactivator 2, mRNA decapping complex subunit 2, mRNA-decapping enzyme subunit 1 | | Authors: | Mugridge, J.S, Ziemniak, M, Jemielity, J, Gross, J.D. | | Deposit date: | 2016-07-05 | | Release date: | 2016-10-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | Structural basis of mRNA-cap recognition by Dcp1-Dcp2.
Nat.Struct.Mol.Biol., 23, 2016
|
|
3FJY
 
 | |
6VCK
 
 | | Crystal structure of E.coli RppH-DapF in complex with GDP, Mg2+ and F- | | Descriptor: | CHLORIDE ION, Diaminopimelate epimerase, FLUORIDE ION, ... | | Authors: | Gao, A, Vasilyev, N, Kaushik, A, Duan, W, Serganov, A. | | Deposit date: | 2019-12-21 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Principles of RNA and nucleotide discrimination by the RNA processing enzyme RppH.
Nucleic Acids Res., 48, 2020
|
|
6VCM
 
 | | Crystal structure of E.coli RppH-DapF in complex with GTP, Mg2+ and F- | | Descriptor: | CHLORIDE ION, Diaminopimelate epimerase, FLUORIDE ION, ... | | Authors: | Gao, A, Vasilyev, N, Kaushik, A, Duan, W, Serganov, A. | | Deposit date: | 2019-12-21 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Principles of RNA and nucleotide discrimination by the RNA processing enzyme RppH.
Nucleic Acids Res., 48, 2020
|
|
6Y3Z
 
 | |
6VCL
 
 | | Crystal structure of E.coli RppH-DapF in complex with pppGpp, Mg2+ and F- | | Descriptor: | CHLORIDE ION, Diaminopimelate epimerase, FLUORIDE ION, ... | | Authors: | Gao, A, Vasilyev, N, Kaushik, A, Duan, W, Serganov, A. | | Deposit date: | 2019-12-21 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Principles of RNA and nucleotide discrimination by the RNA processing enzyme RppH.
Nucleic Acids Res., 48, 2020
|
|
5N2V
 
 | |
5LON
 
 | | Structure of /K. lactis/ Dcp1-Dcp2 decapping complex. | | Descriptor: | KLLA0E01827p, KLLA0F23980p | | Authors: | Charenton, C, Taverniti, V, Gaudon-Plesse, C, Back, R, Seraphin, B, Graille, M. | | Deposit date: | 2016-08-09 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of the active form of Dcp1-Dcp2 decapping enzyme bound to m(7)GDP and its Edc3 activator.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5LOP
 
 | | Structure of the active form of /K. lactis/ Dcp1-Dcp2-Edc3 decapping complex bound to m7GDP | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, KLLA0A11308p, KLLA0E01827p, ... | | Authors: | Charenton, C, Taverniti, V, Gaudon-Plesse, C, Back, R, Seraphin, B, Graille, M. | | Deposit date: | 2016-08-09 | | Release date: | 2016-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of the active form of Dcp1-Dcp2 decapping enzyme bound to m(7)GDP and its Edc3 activator.
Nat.Struct.Mol.Biol., 23, 2016
|
|
3GZ5
 
 | | Crystal structure of Shewanella oneidensis NrtR | | Descriptor: | MutT/nudix family protein, SODIUM ION | | Authors: | Huang, N, Zhang, H. | | Deposit date: | 2009-04-06 | | Release date: | 2009-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and function of an ADP-ribose-dependent transcriptional regulator of NAD metabolism
Structure, 17, 2009
|
|
