3GUL
 
 | |
1MQH
 
 | | Crystal Structure of the GluR2 Ligand Binding Core (S1S2J) in Complex with Bromo-Willardiine at 1.8 Angstroms Resolution | | Descriptor: | 2-AMINO-3-(5-BROMO-2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-PROPIONIC ACID, glutamate receptor 2 | | Authors: | Jin, R, Banke, T.G, Mayer, M.L, Traynelis, S.F, Gouaux, E. | | Deposit date: | 2002-09-16 | | Release date: | 2003-08-05 | | Last modified: | 2017-08-02 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for partial agonist action at ionotropic glutamate receptors
Nat.Neurosci., 6, 2003
|
|
3GUK
 
 | |
6U38
 
 | | PCSK9 in complex with a Fab and compound 8 | | Descriptor: | 2-fluoro-4-{[(1R)-1-methyl-6-{[(2S)-oxan-2-yl]methoxy}-1-{2-oxo-2-[(1,3-thiazol-2-yl)amino]ethyl}-1,2,3,4-tetrahydroisoquinolin-7-yl]oxy}benzoic acid, Fab Heavy Chain, Fab Light Chain, ... | | Authors: | Lu, J, Soisson, S. | | Deposit date: | 2019-08-21 | | Release date: | 2019-11-06 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | From Screening to Targeted Degradation: Strategies for the Discovery and Optimization of Small Molecule Ligands for PCSK9.
Cell Chem Biol, 27, 2020
|
|
6GOH
 
 | | X-ray structure of the adduct formed upon reaction of lysozyme with a Pt(II) complex bearing N,N-pyridylbenzimidazole derivative with an alkylated sulphonate side chain | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, Lysozyme C, ... | | Authors: | Merlino, A, Ferraro, G. | | Deposit date: | 2018-06-01 | | Release date: | 2018-07-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Exploring the interactions between model proteins and Pd(ii) or Pt(ii) compounds bearing charged N,N-pyridylbenzimidazole bidentate ligands by X-ray crystallography.
Dalton Trans, 47, 2018
|
|
6U2N
 
 | | PCSK9 in complex with compound 4 | | Descriptor: | 4-{[(1R)-6-methoxy-1-methyl-1-{2-oxo-2-[(1,3-thiazol-2-yl)amino]ethyl}-1,2,3,4-tetrahydroisoquinolin-7-yl]oxy}benzoic acid, Proprotein convertase subtilisin/kexin type 9 | | Authors: | Lu, J, Soisson, S. | | Deposit date: | 2019-08-20 | | Release date: | 2019-11-06 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | From Screening to Targeted Degradation: Strategies for the Discovery and Optimization of Small Molecule Ligands for PCSK9.
Cell Chem Biol, 27, 2020
|
|
6GOI
 
 | | X-ray structure of the adduct formed upon reaction of lysozyme with a Pd(II) complex bearing N,N-pyridylbenzimidazole derivative with an alkylated triphenylphosphonium cation | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, Lysozyme C, ... | | Authors: | Merlino, A, Ferraro, G. | | Deposit date: | 2018-06-01 | | Release date: | 2018-07-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Exploring the interactions between model proteins and Pd(ii) or Pt(ii) compounds bearing charged N,N-pyridylbenzimidazole bidentate ligands by X-ray crystallography.
Dalton Trans, 47, 2018
|
|
20GS
 
 | | GLUTATHIONE S-TRANSFERASE P1-1 COMPLEXED WITH CIBACRON BLUE | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CIBACRON BLUE, GLUTATHIONE S-TRANSFERASE | | Authors: | Oakley, A.J, Lo Bello, M, Nuccetelli, M, Mazzetti, A.P, Parker, M.W. | | Deposit date: | 1997-12-16 | | Release date: | 1998-12-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The ligandin (non-substrate) binding site of human Pi class glutathione transferase is located in the electrophile binding site (H-site).
J.Mol.Biol., 291, 1999
|
|
6U36
 
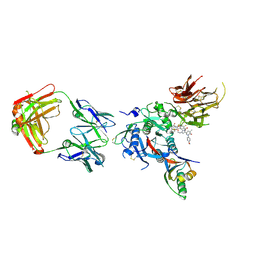 | | PCSK9 in complex with a Fab and compound 14 | | Descriptor: | 2-fluoro-4-{[(1R)-6-(2-{4-[1-(4-methoxyphenyl)-5-methyl-6-oxo-1,6-dihydropyridazin-3-yl]-1H-1,2,3-triazol-1-yl}ethoxy)-1-methyl-1-{2-oxo-2-[(1,3-thiazol-2-yl)amino]ethyl}-1,2,3,4-tetrahydroisoquinolin-7-yl]oxy}benzoic acid, Fab Heavy Chain, Fab Light Chain, ... | | Authors: | Lu, J, Soisson, S. | | Deposit date: | 2019-08-21 | | Release date: | 2019-11-06 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | From Screening to Targeted Degradation: Strategies for the Discovery and Optimization of Small Molecule Ligands for PCSK9.
Cell Chem Biol, 27, 2020
|
|
3FIR
 
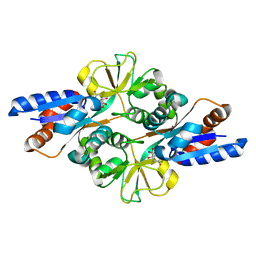 | | Crystal structure of Glycosylated K135E PEB3 | | Descriptor: | 2-acetamido-2-deoxy-alpha-L-glucopyranose-(1-3)-2,4-bisacetamido-2,4,6-trideoxy-beta-D-glucopyranose, CITRATE ANION, Major antigenic peptide PEB3 | | Authors: | Min, T, Matte, A, Cygler, M. | | Deposit date: | 2008-12-12 | | Release date: | 2009-03-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Specificity of Campylobacter jejuni adhesin PEB3 for phosphates and structural differences among its ligand complexes.
Biochemistry, 48, 2009
|
|
8BA4
 
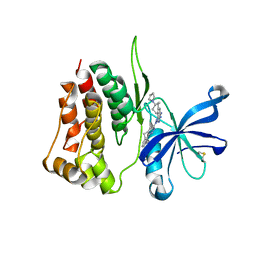 | | Crystal structure of JAK2 JH2-V617F in complex with Bemcentinib | | Descriptor: | 1-(3,4-diazatricyclo[9.4.0.0^{2,7}]pentadeca-1(11),2(7),3,5,12,14-hexaen-5-yl)-~{N}3-[(7~{S})-7-pyrrolidin-1-yl-6,7,8,9-tetrahydro-5~{H}-benzo[7]annulen-3-yl]-1,2,4-triazole-3,5-diamine, Tyrosine-protein kinase JAK2 | | Authors: | Haikarainen, T, Silvennoinen, O. | | Deposit date: | 2022-10-11 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of Novel Small Molecule Ligands for JAK2 Pseudokinase Domain.
Pharmaceuticals, 16, 2023
|
|
8BA3
 
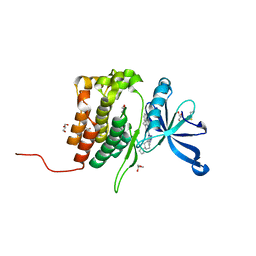 | | Crystal structure of JAK2 JH2 in complex with Bemcentinib | | Descriptor: | 1-(3,4-diazatricyclo[9.4.0.0^{2,7}]pentadeca-1(11),2(7),3,5,12,14-hexaen-5-yl)-~{N}3-[(7~{S})-7-pyrrolidin-1-yl-6,7,8,9-tetrahydro-5~{H}-benzo[7]annulen-3-yl]-1,2,4-triazole-3,5-diamine, GLYCEROL, Tyrosine-protein kinase JAK2 | | Authors: | Haikarainen, T, Silvennoinen, O. | | Deposit date: | 2022-10-11 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Identification of Novel Small Molecule Ligands for JAK2 Pseudokinase Domain.
Pharmaceuticals, 16, 2023
|
|
8B99
 
 | | Crystal structure of JAK2 JH2-V617F in complex with JNJ-7706621 | | Descriptor: | 4-({5-amino-1-[(2,6-difluorophenyl)carbonyl]-1H-1,2,4-triazol-3-yl}amino)benzenesulfonamide, GLYCEROL, Tyrosine-protein kinase JAK2 | | Authors: | Haikarainen, T, Silvennoinen, O. | | Deposit date: | 2022-10-05 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification of Novel Small Molecule Ligands for JAK2 Pseudokinase Domain.
Pharmaceuticals, 16, 2023
|
|
8B8U
 
 | |
8B9H
 
 | | Crystal structure of JAK2 JH2 in complex with Z902-A3 | | Descriptor: | 6-[[methyl-[(3-methylthiophen-2-yl)methyl]amino]methyl]-~{N}4-phenyl-1,3,5-triazine-2,4-diamine, GLYCEROL, Tyrosine-protein kinase JAK2 | | Authors: | Haikarainen, T, Silvennoinen, O. | | Deposit date: | 2022-10-06 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Identification of Novel Small Molecule Ligands for JAK2 Pseudokinase Domain.
Pharmaceuticals, 16, 2023
|
|
8B9E
 
 | | Crystal structure of JAK2 JH2-V617F in complex with Z902-A3 | | Descriptor: | 6-[[methyl-[(3-methylthiophen-2-yl)methyl]amino]methyl]-~{N}4-phenyl-1,3,5-triazine-2,4-diamine, GLYCEROL, Tyrosine-protein kinase JAK2 | | Authors: | Haikarainen, T, Silvennoinen, O. | | Deposit date: | 2022-10-05 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Identification of Novel Small Molecule Ligands for JAK2 Pseudokinase Domain.
Pharmaceuticals, 16, 2023
|
|
8BAK
 
 | |
8BAB
 
 | | Crystal structure of JAK2 JH2-V617F in complex with CB76 | | Descriptor: | 6-[(1-methylimidazol-2-yl)sulfanylmethyl]-~{N}4-(3-methylphenyl)-1,3,5-triazine-2,4-diamine, GLYCEROL, Tyrosine-protein kinase JAK2 | | Authors: | Haikarainen, T, Silvennoinen, O. | | Deposit date: | 2022-10-11 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Identification of Novel Small Molecule Ligands for JAK2 Pseudokinase Domain.
Pharmaceuticals, 16, 2023
|
|
8B8N
 
 | |
8BA2
 
 | | Crystal structure of JAK2 JH2-V617F in complex with Z902-A1 | | Descriptor: | 6-[[(5-bromanylthiophen-2-yl)methyl-methyl-amino]methyl]-~{N}4-(4-methylphenyl)-1,3,5-triazine-2,4-diamine, ACETATE ION, GLYCEROL, ... | | Authors: | Haikarainen, T, Silvennoinen, O. | | Deposit date: | 2022-10-11 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Identification of Novel Small Molecule Ligands for JAK2 Pseudokinase Domain.
Pharmaceuticals, 16, 2023
|
|
1YQ9
 
 | | Structure of the unready oxidized form of [NiFe] hydrogenase | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, GLYCEROL, ... | | Authors: | Volbeda, A, Martin, L, Cavazza, C, Matho, M, Faber, B.W, Roseboom, W, Albracht, S.P, Garcin, E, Rousset, M, Fontecilla-Camps, J.C. | | Deposit date: | 2005-02-01 | | Release date: | 2005-04-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural differences between the ready and unready oxidized states of [NiFe] hydrogenases.
J.Biol.Inorg.Chem., 10, 2005
|
|
6U3X
 
 | | PCSK9 in complex with compound 2 | | Descriptor: | 2-[(1R)-6,7-dimethoxy-1-methyl-1,2,3,4-tetrahydroisoquinolin-1-yl]-N-(1,3-thiazol-2-yl)acetamide, Proprotein convertase subtilisin/kexin type 9 | | Authors: | Lu, J, Soisson, S. | | Deposit date: | 2019-08-22 | | Release date: | 2019-11-06 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | From Screening to Targeted Degradation: Strategies for the Discovery and Optimization of Small Molecule Ligands for PCSK9.
Cell Chem Biol, 27, 2020
|
|
6U26
 
 | | PCSK9 in complex with compound 16 | | Descriptor: | 4'-{[(1R)-6-{2-[2-({N~5~-[N,N'-bis(tert-butoxycarbonyl)carbamimidoyl]-N~2~-(tert-butoxycarbonyl)-L-ornithyl}amino)ethoxy]ethoxy}-1-methyl-1-{2-oxo-2-[(1,3-thiazol-2-yl)amino]ethyl}-1,2,3,4-tetrahydroisoquinolin-7-yl]oxy}-2'-fluoro[1,1'-biphenyl]-4-carboxylic acid, Proprotein convertase subtilisin/kexin type 9 | | Authors: | Orth, P. | | Deposit date: | 2019-08-19 | | Release date: | 2019-11-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | From Screening to Targeted Degradation: Strategies for the Discovery and Optimization of Small Molecule Ligands for PCSK9.
Cell Chem Biol, 27, 2020
|
|
6U2P
 
 | | PCSK9 in complex with compound 5 | | Descriptor: | 1,2-ETHANEDIOL, 2-fluoro-4-{[(1R)-6-methoxy-1-methyl-1-{2-oxo-2-[(1,3-thiazol-2-yl)amino]ethyl}-1,2,3,4-tetrahydroisoquinolin-7-yl]oxy}benzoic acid, Proprotein convertase subtilisin/kexin type 9 | | Authors: | Lu, J, Soisson, S. | | Deposit date: | 2019-08-20 | | Release date: | 2019-11-06 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | From Screening to Targeted Degradation: Strategies for the Discovery and Optimization of Small Molecule Ligands for PCSK9.
Cell Chem Biol, 27, 2020
|
|
6E2B
 
 | | Ubiquitin in complex with Pt(2-phenilpyridine)(PPh3) | | Descriptor: | GLYCEROL, SULFATE ION, Ubiquitin, ... | | Authors: | Zhemkov, V.A, Kim, M. | | Deposit date: | 2018-07-11 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Reactions of Cyclometalated Platinum(II) [Pt(N∧C)(PR3)Cl] Complexes with Imidazole and Imidazole-Containing Biomolecules: Fine-Tuning of Reactivity and Photophysical Properties via Ligand Design.
Inorg Chem, 58, 2019
|
|
