1A0N
 
 | | NMR STUDY OF THE SH3 DOMAIN FROM FYN PROTO-ONCOGENE TYROSINE KINASE COMPLEXED WITH THE SYNTHETIC PEPTIDE P2L CORRESPONDING TO RESIDUES 91-104 OF THE P85 SUBUNIT OF PI3-KINASE, FAMILY OF 25 STRUCTURES | | Descriptor: | FYN, PRO-PRO-ARG-PRO-LEU-PRO-VAL-ALA-PRO-GLY-SER-SER-LYS-THR | | Authors: | Renzoni, D.A, Pugh, D.J.R, Siligardi, G, Das, P, Morton, C.J, Rossi, C, Waterfield, M.D, Campbell, I.D, Ladbury, J.E. | | Deposit date: | 1997-12-05 | | Release date: | 1998-02-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural and thermodynamic characterization of the interaction of the SH3 domain from Fyn with the proline-rich binding site on the p85 subunit of PI3-kinase.
Biochemistry, 35, 1996
|
|
1AOT
 
 | | NMR STRUCTURE OF THE FYN SH2 DOMAIN COMPLEXED WITH A PHOSPHOTYROSYL PEPTIDE, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | FYN PROTEIN-TYROSINE KINASE, PHOSPHOTYROSYL PEPTIDE | | Authors: | Mulhern, T.D, Shaw, G.L, Morton, C.J, Day, A.J, Campbell, I.D. | | Deposit date: | 1997-07-10 | | Release date: | 1998-01-14 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | The SH2 domain from the tyrosine kinase Fyn in complex with a phosphotyrosyl peptide reveals insights into domain stability and binding specificity.
Structure, 5, 1997
|
|
1AOU
 
 | | NMR STRUCTURE OF THE FYN SH2 DOMAIN COMPLEXED WITH A PHOSPHOTYROSYL PEPTIDE, 22 STRUCTURES | | Descriptor: | FYN PROTEIN-TYROSINE KINASE, PHOSPHOTYROSYL PEPTIDE | | Authors: | Mulhern, T.D, Shaw, G.L, Morton, C.J, Day, A.J, Campbell, I.D. | | Deposit date: | 1997-07-10 | | Release date: | 1998-01-14 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | The SH2 domain from the tyrosine kinase Fyn in complex with a phosphotyrosyl peptide reveals insights into domain stability and binding specificity.
Structure, 5, 1997
|
|
1K2J
 
 | | NMR MINIMIZED AVERAGE STRUCTURE OF d(CGTACG)2 | | Descriptor: | 5'-D(*CP*GP*TP*AP*CP*G)-3' | | Authors: | Lam, S.L, Ip, L.N. | | Deposit date: | 2001-09-27 | | Release date: | 2002-04-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Low temperature solution structures and base pair stacking of double helical d(CGTACG)(2).
J.Biomol.Struct.Dyn., 19, 2002
|
|
1AZG
 
 | | NMR STUDY OF THE SH3 DOMAIN FROM FYN PROTO-ONCOGENE TYROSINE KINASE KINASE COMPLEXED WITH THE SYNTHETIC PEPTIDE P2L CORRESPONDING TO RESIDUES 91-104 OF THE P85 SUBUNIT OF PI3-KINASE, MINIMIZED AVERAGE (PROBMAP) STRUCTURE | | Descriptor: | FYN, PRO-PRO-ARG-PRO-LEU-PRO-VAL-ALA-PRO-GLY-SER-SER-LYS-THR | | Authors: | Renzoni, D.A, Pugh, D.J.R, Siligardi, G, Das, P, Morton, C.J, Rossi, C, Waterfield, M.D, Campbell, I.D, Ladbury, J.E. | | Deposit date: | 1997-11-18 | | Release date: | 1998-02-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural and thermodynamic characterization of the interaction of the SH3 domain from Fyn with the proline-rich binding site on the p85 subunit of PI3-kinase.
Biochemistry, 35, 1996
|
|
6TV5
 
 | |
1K2K
 
 | | NMR MINIMIZED AVERAGE STRUCTURE OF d(CGTACG)2 | | Descriptor: | 5'-D(*CP*GP*TP*AP*CP*G)-3' | | Authors: | Lam, S.L, Ip, L.N. | | Deposit date: | 2001-09-28 | | Release date: | 2002-04-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Low temperature solution structures and base pair stacking of double helical d(CGTACG)(2).
J.Biomol.Struct.Dyn., 19, 2002
|
|
2KEZ
 
 | | NMR structure of U6 ISL at pH 8.0 | | Descriptor: | RNA (5'-R(*GP*GP*UP*UP*CP*CP*CP*CP*UP*GP*CP*AP*UP*AP*AP*GP*GP*AP*UP*GP*AP*AP*CP*C)-3') | | Authors: | Venditti, V, Butcher, S.E. | | Deposit date: | 2009-02-08 | | Release date: | 2009-07-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Minimum-energy path for a u6 RNA conformational change involving protonation, base-pair rearrangement and base flipping.
J.Mol.Biol., 391, 2009
|
|
1HJ7
 
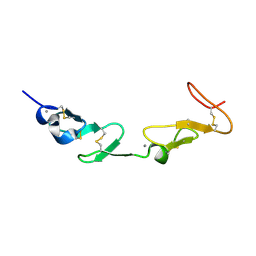 | | NMR study of a pair of LDL receptor Ca2+ binding epidermal growth factor-like domains, 20 structures | | Descriptor: | CALCIUM ION, LDL RECEPTOR | | Authors: | Saha, S, Handford, P.A, Campbell, I.D, Downing, A.K. | | Deposit date: | 2001-01-09 | | Release date: | 2001-07-11 | | Last modified: | 2018-02-14 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Ldl Receptor Egf-Ab Pair: A Paradigm for the Assembly of Tandem Calcium Binding Egf Domains
Structure, 9, 2001
|
|
1SM7
 
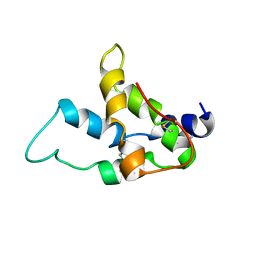 | | Solution structure of the recombinant pronapin precursor, BnIb. | | Descriptor: | recombinant Ib pronapin | | Authors: | Pantoja-Uceda, D, Palomares, O, Bruix, M, Villalba, M, Rodriguez, R, Rico, M, Santoro, J. | | Deposit date: | 2004-03-08 | | Release date: | 2005-02-01 | | Last modified: | 2013-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure and stability against digestion of rproBnIb, a recombinant 2S albumin from rapeseed: relationship to its allergenic properties.
Biochemistry, 43, 2004
|
|
2KDC
 
 | | NMR Solution Structure of E. coli diacylglycerol kinase (DAGK) in DPC micelles | | Descriptor: | Diacylglycerol kinase | | Authors: | Van Horn, W.D, Kim, H, Ellis, C.D, Hadziselimovic, A, Sulistijo, E.S, Karra, M.D, Tian, C, Sonnichsen, F.D, Sanders, C.R. | | Deposit date: | 2009-01-06 | | Release date: | 2009-07-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution nuclear magnetic resonance structure of membrane-integral diacylglycerol kinase
Science, 324, 2009
|
|
1ROO
 
 | | NMR SOLUTION STRUCTURE OF SHK TOXIN, NMR, 20 STRUCTURES | | Descriptor: | SHK TOXIN | | Authors: | Tudor, J.E, Pallaghy, P.K, Pennington, M.W, Norton, R.S. | | Deposit date: | 1996-01-11 | | Release date: | 1997-01-27 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of ShK toxin, a novel potassium channel inhibitor from a sea anemone.
Nat.Struct.Biol., 3, 1996
|
|
1HFF
 
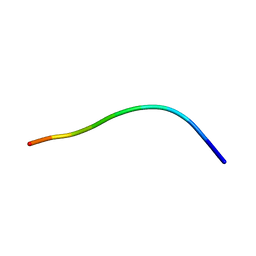 | | NMR solution structures of the vMIP-II 1-10 peptide from Kaposi's sarcoma-associated herpesvirus. | | Descriptor: | VIRAL MACROPHAGE INFLAMMATORY PROTEIN-II | | Authors: | Crump, M.P, Elisseeva, E, Gong, J.H, Clark-Lewis, I, Sykes, B.D. | | Deposit date: | 2000-12-01 | | Release date: | 2000-12-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure/Function of Human Herpesvirus-8 Mip-II (1-71) and the Antagonist N-Terminal Segment (1-10)
FEBS Lett., 489, 2001
|
|
4BS2
 
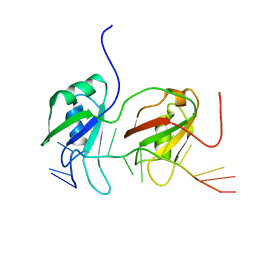 | | NMR structure of human TDP-43 tandem RRMs in complex with UG-rich RNA | | Descriptor: | 5'-R(*GP*UP*GP*UP*GP*AP*AP*UP*GP*AP*AP*UP)-3', TAR DNA-BINDING PROTEIN 43 | | Authors: | Lukavsky, P.J, Daujotyte, D, Tollervey, J.R, Ule, J, Stuani, C, Buratti, E, Baralle, F.E, Damberger, F.F, Allain, F.H.T. | | Deposit date: | 2013-06-06 | | Release date: | 2013-11-13 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Molecular Basis of Ug-Rich RNA Recognition by the Human Splicing Factor Tdp-43
Nat.Struct.Mol.Biol., 20, 2013
|
|
1BI6
 
 | | NMR STRUCTURE OF BROMELAIN INHIBITOR VI FROM PINEAPPLE STEM | | Descriptor: | BROMELAIN INHIBITOR VI | | Authors: | Hatano, K.-I. | | Deposit date: | 1995-12-07 | | Release date: | 1996-04-03 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of bromelain inhibitor IV from pineapple stem: structural similarity with Bowman-Birk trypsin/chymotrypsin inhibitor from soybean.
Biochemistry, 35, 1996
|
|
2Z2G
 
 | | NMR Structure of the IQ-modified Dodecamer CTC[IQ]GGCGCCATC | | Descriptor: | 3-METHYL-3H-IMIDAZO[4,5-F]QUINOLIN-2-AMINE, DNA (5'-D(*DCP*DTP*DCP*DGP*DGP*DCP*DGP*DCP*DCP*DAP*DTP*DC)-3'), DNA (5'-D(*DGP*DAP*DTP*DGP*DGP*DCP*DGP*DCP*DCP*DGP*DAP*DG)-3') | | Authors: | Wang, F, Elmquist, C.E, Stover, J.S, Rizzo, C.J, Stone, M.P. | | Deposit date: | 2007-05-22 | | Release date: | 2007-10-02 | | Last modified: | 2023-11-29 | | Method: | SOLUTION NMR | | Cite: | DNA sequence modulates the conformation of the food mutagen 2-amino-3-methylimidazo[4,5-f]quinoline in the recognition sequence of the NarI restriction enzyme
Biochemistry, 46, 2007
|
|
2Z2H
 
 | | NMR Structure of the IQ-modified Dodecamer CTCG[IQ]GCGCCATC | | Descriptor: | 3-METHYL-3H-IMIDAZO[4,5-F]QUINOLIN-2-AMINE, DNA (5'-D(*DCP*DTP*DCP*DGP*DGP*DCP*DGP*DCP*DCP*DAP*DTP*DC)-3'), DNA (5'-D(*DGP*DAP*DTP*DGP*DGP*DCP*DGP*DCP*DCP*DGP*DAP*DG)-3') | | Authors: | Wang, F, Elmquist, C.E, Stover, J.S, Rizzo, C.J, Stone, M.P. | | Deposit date: | 2007-05-22 | | Release date: | 2007-10-02 | | Last modified: | 2023-11-29 | | Method: | SOLUTION NMR | | Cite: | DNA sequence modulates the conformation of the food mutagen 2-amino-3-methylimidazo[4,5-f]quinoline in the recognition sequence of the NarI restriction enzyme
Biochemistry, 46, 2007
|
|
1CJG
 
 | | NMR STRUCTURE OF LAC REPRESSOR HP62-DNA COMPLEX | | Descriptor: | DNA (5'-D(*GP*AP*AP*TP*TP*GP*TP*GP*AP*GP*CP*GP*CP*TP*CP*AP*CP*AP*AP*TP*TP*C)-3'), PROTEIN (LAC REPRESSOR) | | Authors: | Spronk, C.A.E.M, Bonvin, A.M.J.J, Radha, P.K, Melacini, G, Boelens, R, Kaptein, R. | | Deposit date: | 1999-04-14 | | Release date: | 2000-01-01 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The solution structure of Lac repressor headpiece 62 complexed to a symmetrical lac operator.
Structure Fold.Des., 7, 1999
|
|
1SKP
 
 | | NMR STRUCTURE OF D(GCATATGATAG)(DOT)D(CTATCATATGC): A CONSENSUS SEQUENCE FOR PROMOTERS RECOGNIZED BY SIGMA-K RNA POLYMERASE, 4 STRUCTURES | | Descriptor: | SIGMA-K RNA POLYMERASE CONSENSUS SEQUENCE | | Authors: | Tonelli, M, Ragg, E, Bianucci, A.M, Lesiak, K, James, T.L. | | Deposit date: | 1998-05-20 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structure of d(GCATATGATAG). d(CTATCATATGC): a consensus sequence for promoters recognized by sigma K RNA polymerase.
Biochemistry, 37, 1998
|
|
1UAB
 
 | | NMR structure of hemimethylated GATC site | | Descriptor: | DNA (5'-D(*CP*GP*CP*AP*GP*(6MA)P*TP*CP*TP*CP*GP*C)-3'), DNA (5'-D(*GP*CP*GP*AP*GP*AP*TP*CP*TP*GP*CP*G)-3') | | Authors: | Bae, S.-H, Cheong, H.-K, Kang, S, Hwang, D.S, Cheong, C, Choi, B.-S. | | Deposit date: | 2003-03-07 | | Release date: | 2004-04-27 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of hemimethylated GATC sites: implications for DNA-SeqA recognition
J.Biol.Chem., 278, 2003
|
|
1NGU
 
 | | NMR Structure of Putative 3'Terminator for B. Anthracis pagA Gene Noncoding Strand | | Descriptor: | 5'-D(*CP*TP*CP*TP*CP*CP*TP*TP*GP*TP*AP*TP*TP*TP*CP*TP*TP*AP*CP*AP*AP*AP*AP*AP*GP*AP*G)-3' | | Authors: | Shiflett, P.R, Taylor-McCabe, K.J, Michalczyk, R, Silks, L.A, Gupta, G. | | Deposit date: | 2002-12-17 | | Release date: | 2003-06-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Studies on the Hairpins at the 3' Untranslated Region of an Anthrax Toxin Gene
Biochemistry, 42, 2003
|
|
1IMO
 
 | |
1HFG
 
 | | NMR solution structure of vMIP-II 1-71 from Kaposi's sarcoma-associated herpesvirus (minimized average structure). | | Descriptor: | VIRAL MACROPHAGE INFLAMMATORY PROTEIN-II | | Authors: | Crump, M.P, Elisseeva, E, Gong, J.-H, Clark-Lewis, I, Sykes, B.D. | | Deposit date: | 2000-12-01 | | Release date: | 2001-01-07 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structure/Function of Human Herpesvirus-8 Mip-II (1-71) and the Antagonist N-Terminal Segment (1-10)
FEBS Lett., 489, 2001
|
|
1IBX
 
 | | NMR STRUCTURE OF DFF40 AND DFF45 N-TERMINAL DOMAIN COMPLEX | | Descriptor: | CHIMERA OF IGG BINDING PROTEIN G AND DNA FRAGMENTATION FACTOR 45, DNA FRAGMENTATION FACTOR 40 | | Authors: | Zhou, P, Lugovskoy, A.A, McCarty, J.S, Li, P, Wagner, G. | | Deposit date: | 2001-03-29 | | Release date: | 2001-05-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of DFF40 and DFF45 N-terminal domain complex and mutual chaperone activity of DFF40 and DFF45.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1OQ2
 
 | | NMR structure of hemimethylated GATC site | | Descriptor: | 5'-D(*CP*GP*CP*AP*GP*(6MA)P*TP*CP*TP*CP*GP*C)-3', 5'-D(*GP*CP*GP*AP*GP*AP*TP*CP*TP*GP*CP*G)-3' | | Authors: | Bae, S.-H, Cheong, H.-K, Kang, S, Hwang, D.S, Cheong, C, Choi, B.-S. | | Deposit date: | 2003-03-07 | | Release date: | 2004-04-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of hemimethylated GATC sites: implications for DNA-SeqA recognition
J.Biol.Chem., 278, 2003
|
|
