1WOG
 
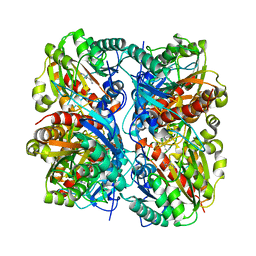 | | Crystal Structure of Agmatinase Reveals Structural Conservation and Inhibition Mechanism of the Ureohydrolase Superfamily | | Descriptor: | HEXANE-1,6-DIAMINE, MANGANESE (II) ION, agmatinase | | Authors: | Ahn, H.J, Kim, K.H, Lee, J, Ha, J.-Y, Lee, H.H, Kim, D, Yoon, H.-J, Kwon, A.-R, Suh, S.W. | | Deposit date: | 2004-08-18 | | Release date: | 2004-09-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of agmatinase reveals structural conservation and inhibition mechanism of the ureohydrolase superfamily
J.Biol.Chem., 279, 2004
|
|
4TGL
 
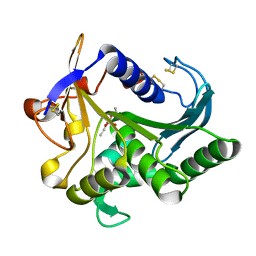 | | CATALYSIS AT THE INTERFACE: THE ANATOMY OF A CONFORMATIONAL CHANGE IN A TRIGLYCERIDE LIPASE | | Descriptor: | DIETHYL PHOSPHONATE, TRIACYL-GLYCEROL ACYLHYDROLASE | | Authors: | Derewenda, U, Brzozowski, A.M, Lawson, D, Derewenda, Z.S. | | Deposit date: | 1991-07-29 | | Release date: | 1993-07-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Catalysis at the interface: the anatomy of a conformational change in a triglyceride lipase.
Biochemistry, 31, 1992
|
|
1WOH
 
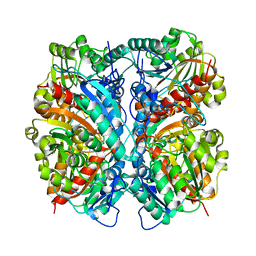 | | Crystal Structure of Agmatinase Reveals Structural Conservation and Inhibition Mechanism of the Ureohydrolase Superfamily | | Descriptor: | agmatinase | | Authors: | Ahn, H.J, Kim, K.H, Lee, J, Ha, J.-Y, Lee, H.H, Kim, D, Yoon, H.-J, Kwon, A.-R, Suh, S.W. | | Deposit date: | 2004-08-18 | | Release date: | 2004-09-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of agmatinase reveals structural conservation and inhibition mechanism of the ureohydrolase superfamily
J.Biol.Chem., 279, 2004
|
|
1WOI
 
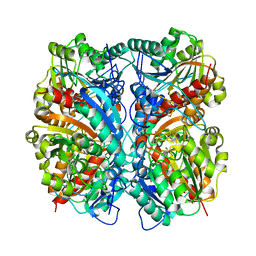 | | Crystal Structure of Agmatinase Reveals Structural Conservation and Inhibition Mechanism of the Ureohydrolase Superfamily | | Descriptor: | MANGANESE (II) ION, agmatinase | | Authors: | Ahn, H.J, Kim, K.H, Lee, J, Ha, J.-Y, Lee, H.H, Kim, D, Yoon, H.-J, Kwon, A.-R, Suh, S.W. | | Deposit date: | 2004-08-18 | | Release date: | 2004-09-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of agmatinase reveals structural conservation and inhibition mechanism of the ureohydrolase superfamily
J.Biol.Chem., 279, 2004
|
|
8DFH
 
 | |
8DFI
 
 | |
2RPJ
 
 | | Solution structure of Fn14 CRD domain | | Descriptor: | Tumor necrosis factor receptor superfamily member 12A | | Authors: | He, F, Dang, W, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-05-19 | | Release date: | 2009-03-24 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the cysteine-rich domain in Fn14, a member of the tumor necrosis factor receptor superfamily
Protein Sci., 18, 2009
|
|
8D2Z
 
 | |
5MP6
 
 | | Structure of the Unliganded Fab from HIV-1 Neutralizing Antibody CAP248-2B that Binds to the gp120 C-terminus - gp41 Interface, at two Angstrom resolution. | | Descriptor: | CAP248-2B Heavy Chain, CAP248-2B Light Chain, SULFATE ION | | Authors: | Wibmer, C.K, Gorman, J, Kwong, P.D. | | Deposit date: | 2016-12-15 | | Release date: | 2016-12-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.959 Å) | | Cite: | Structure and Recognition of a Novel HIV-1 gp120-gp41 Interface Antibody that Caused MPER Exposure through Viral Escape.
PLoS Pathog., 13, 2017
|
|
4B4N
 
 | |
3LDA
 
 | | Yeast Rad51 H352Y Filament Interface Mutant | | Descriptor: | CHLORIDE ION, DNA repair protein RAD51 | | Authors: | Villanueva, N.L, Chen, J, Morrical, S.W, Rould, M.A. | | Deposit date: | 2010-01-12 | | Release date: | 2010-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights into the mechanism of Rad51 recombinase from the structure and properties of a filament interface mutant.
Nucleic Acids Res., 38, 2010
|
|
1PX5
 
 | | Crystal structure of the 2'-specific and double-stranded RNA-activated interferon-induced antiviral protein 2'-5'-oligoadenylate synthetase | | Descriptor: | 2'-5'-oligoadenylate synthetase 1, SULFATE ION | | Authors: | Hartmann, R, Justesen, J, Sarkar, S.N, Sen, G.C, Yee, V.C. | | Deposit date: | 2003-07-02 | | Release date: | 2003-11-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structure of the 2'-specific and double-stranded RNA-activated interferon-induced antiviral protein 2'-5'-oligoadenylate synthetase
Mol.Cell, 12, 2003
|
|
3IQ6
 
 | |
1E7Z
 
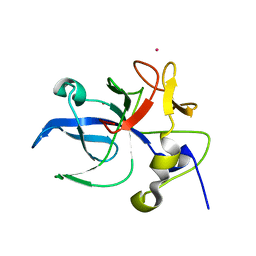 | | Crystal structure of the EMAP2/RNA binding domain of the p43 protein from human aminoacyl-tRNA synthetase complex | | Descriptor: | ENDOTHELIAL-MONOCYTE ACTIVATING POLYPEPTIDE II, MERCURY (II) ION | | Authors: | Pasqualato, S, Kerjan, P, Renault, L, Menetrey, J, Mirande, M, Cherfils, J. | | Deposit date: | 2000-09-13 | | Release date: | 2000-11-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of the Emapii Domain of Human Aminoacyl-tRNA Synthetase Complex Reveals Evolutionary Dimeric Mimicry
Embo J., 20, 2001
|
|
2F3R
 
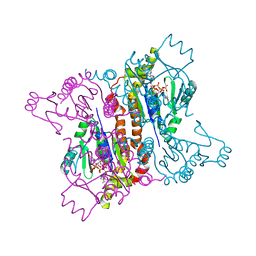 | |
2F3T
 
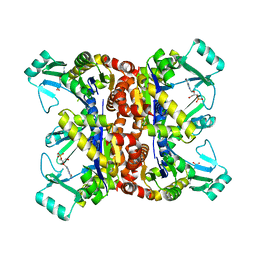 | |
3VOW
 
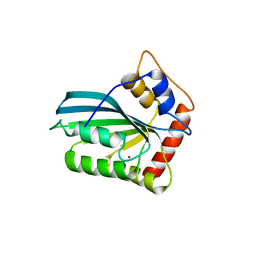 | | Crystal Structure of the Human APOBEC3C having HIV-1 Vif-binding Interface | | Descriptor: | CHLORIDE ION, Probable DNA dC->dU-editing enzyme APOBEC-3C, ZINC ION | | Authors: | Kitamura, S, Suzuki, A, Watanabe, N, Iwatani, Y. | | Deposit date: | 2012-02-22 | | Release date: | 2012-10-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The APOBEC3C crystal structure and the interface for HIV-1 Vif binding.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3CXH
 
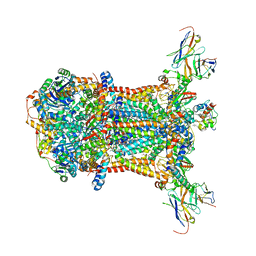 | |
8OYE
 
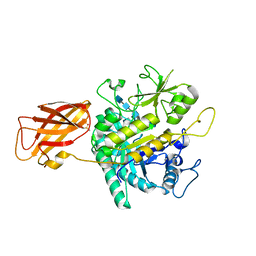 | | Clostridium perfringens chitinase CP4_3455 E196Q with chitin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitodextrinase, DIMETHYL SULFOXIDE | | Authors: | Bloch, Y, Savvides, S.N. | | Deposit date: | 2023-05-04 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Clostridium perfringens chitinase CP4_3455 E196Q with chitin
To Be Published
|
|
8OSE
 
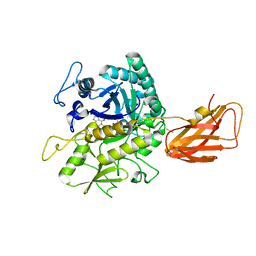 | | C. perfringens chitinase CP4_3455 in complex with inhibitor bisdionin C | | Descriptor: | 1,1'-PROPANE-1,3-DIYLBIS(3,7-DIMETHYL-3,7-DIHYDRO-1H-PURINE-2,6-DIONE), 1,2-ETHANEDIOL, Chitodextrinase, ... | | Authors: | Bloch, Y, Savvides, S.N. | | Deposit date: | 2023-04-18 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | C. perfringens chitinase CP4_3455 in complex with inhibitor bisdionin C
To Be Published
|
|
8OTB
 
 | |
1FL0
 
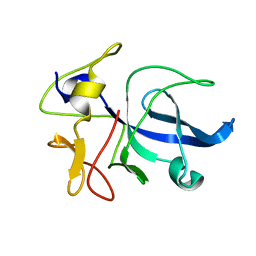 | | CRYSTAL STRUCTURE OF THE EMAP2/RNA-BINDING DOMAIN OF THE P43 PROTEIN FROM HUMAN AMINOACYL-TRNA SYNTHETASE COMPLEX | | Descriptor: | ENDOTHELIAL-MONOCYTE ACTIVATING POLYPEPTIDE II | | Authors: | Renault, L, Kerjan, P, Pasqualato, S, Menetrey, J, Robinson, J.-C, Kawaguchi, S, Vassylyev, D.G, Yokoyama, S, Mirande, M, Cherfils, J. | | Deposit date: | 2000-08-11 | | Release date: | 2000-12-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the EMAPII domain of human aminoacyl-tRNA synthetase complex reveals evolutionary dimer mimicry.
EMBO J., 20, 2001
|
|
3M79
 
 | |
3C3S
 
 | | Role of a Glutamate Bridge Spanning the Dimeric Interface of Human Manganese Superoxide Dismutase | | Descriptor: | MANGANESE (II) ION, SULFATE ION, Superoxide dismutase [Mn] | | Authors: | Quint, P.S, Domsic, J.F, Cabelli, D.E, McKenna, R, Silverman, D.N. | | Deposit date: | 2008-01-28 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Role of a glutamate bridge spanning the dimeric interface of human manganese superoxide dismutase.
Biochemistry, 47, 2008
|
|
3SXU
 
 | |
