8IKW
 
 | | A complex structure of PGIP-PG | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Endo-polygalacturonase, ... | | Authors: | Xiao, Y, Chai, J. | | Deposit date: | 2023-03-01 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | A plant mechanism of hijacking pathogen virulence factors to trigger innate immunity.
Science, 383, 2024
|
|
3WH0
 
 | | Structure of Pin1 Complex with 18-crown-6 | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, ... | | Authors: | Lee, C.C, Liu, C.I, Jeng, W.Y, Wang, A.H.J. | | Deposit date: | 2013-08-20 | | Release date: | 2014-10-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crowning proteins: modulating the protein surface properties using crown ethers.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
5EXD
 
 | | Crystal structure of oxalate oxidoreductase from Moorella thermoacetica bound with carboxy-di-oxido-methyl-TPP (COOM-TPP) intermediate | | Descriptor: | IRON/SULFUR CLUSTER, MAGNESIUM ION, Oxalate oxidoreductase subunit alpha, ... | | Authors: | Gibson, M.I, Chen, P.Y.-T, Drennan, C.L. | | Deposit date: | 2015-11-23 | | Release date: | 2015-12-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | One-carbon chemistry of oxalate oxidoreductase captured by X-ray crystallography.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7WMV
 
 | | Structure of human SGLT1-MAP17 complex bound with LX2761 | | Descriptor: | N-[2-(dimethylamino)ethyl]-2-methyl-2-[4-[4-[[2-methyl-5-[(2S,3R,4R,5S,6R)-6-methylsulfanyl-3,4,5-tris(oxidanyl)oxan-2-yl]phenyl]methyl]phenyl]butanoylamino]propanamide, PDZK1-interacting protein 1, Sodium/glucose cotransporter 1 | | Authors: | Chen, L, Niu, Y, Cui, W. | | Deposit date: | 2022-01-17 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural mechanism of SGLT1 inhibitors.
Nat Commun, 13, 2022
|
|
5ZCK
 
 | | Structure of the RIP3 core region | | Descriptor: | SODIUM ION, peptide from Receptor-interacting serine/threonine-protein kinase 3 | | Authors: | Li, J, Wu, H. | | Deposit date: | 2018-02-18 | | Release date: | 2018-04-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.271 Å) | | Cite: | The Structure of the Necrosome RIPK1-RIPK3 Core, a Human Hetero-Amyloid Signaling Complex.
Cell, 173, 2018
|
|
2Q81
 
 | | Crystal Structure of the Miz-1 BTB/POZ domain | | Descriptor: | Miz-1 protein, TETRAETHYLENE GLYCOL | | Authors: | Stead, M.A, Trinh, C.H, Garnett, J.A, Carr, S.B, Edwards, T.A, Wright, S.C. | | Deposit date: | 2007-06-08 | | Release date: | 2007-11-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Beta-Sheet Interaction Interface Directs the Tetramerisation of the Miz-1 POZ Domain
J.Mol.Biol., 373, 2007
|
|
3VHT
 
 | | Crystal structure of GFP-Wrnip1 UBZ domain fusion protein in complex with ubiquitin | | Descriptor: | Green fluorescent protein, Green fluorescent protein,ATPase WRNIP1, Ubiquitin, ... | | Authors: | Suzuki, N, Wakatsuki, S, Kawasaki, M. | | Deposit date: | 2011-09-06 | | Release date: | 2012-10-10 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A novel mode of ubiquitin recognition by the ubiquitin-binding zinc finger domain of WRNIP1.
Febs J., 2016
|
|
3ZEH
 
 | |
4ROJ
 
 | | Crystal Structure of the VAV2 SH2 domain in complex with TXNIP phosphorylated peptide | | Descriptor: | Guanine nucleotide exchange factor VAV2, Thioredoxin-interacting protein, UNKNOWN ATOM OR ION | | Authors: | Liu, Y, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-10-28 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of the VAV2 SH2 domain in complex with TXNIP phosphorylated peptide
TO BE PUBLISHED
|
|
3VHS
 
 | |
4BJ6
 
 | | Crystal structure Rif2 in complex with the C-terminal domain of Rap1 (Rap1-RCT) | | Descriptor: | DNA-BINDING PROTEIN RAP1, RAP1-INTERACTING FACTOR 2, SULFATE ION | | Authors: | Shi, T, Bunker, R.D, Gut, H, Scrima, A, Thoma, N.H. | | Deposit date: | 2013-04-16 | | Release date: | 2013-06-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.26 Å) | | Cite: | Rif1 and Rif2 Shape Telomere Funcation and Architecture Through Multivalent RAP1 Interactions
Cell(Cambridge,Mass.), 153, 2013
|
|
8WQ9
 
 | |
7CEI
 
 | | THE ENDONUCLEASE DOMAIN OF COLICIN E7 IN COMPLEX WITH ITS INHIBITOR IM7 PROTEIN | | Descriptor: | PROTEIN (COLICIN E7 IMMUNITY PROTEIN), ZINC ION | | Authors: | Ko, T.P, Liao, C.C, Ku, W.Y, Chak, K.F, Yuan, H.S. | | Deposit date: | 1998-09-17 | | Release date: | 1999-09-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of the DNase domain of colicin E7 in complex with its inhibitor Im7 protein.
Structure Fold.Des., 7, 1999
|
|
4TS6
 
 | |
6M62
 
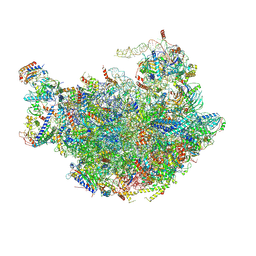 | |
2WWZ
 
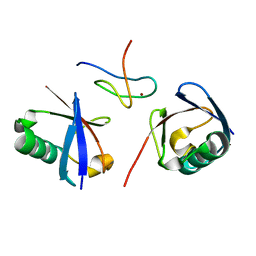 | | TAB2 NZF DOMAIN IN COMPLEX WITH Lys63-linked di-ubiquitin, P212121 | | Descriptor: | MITOGEN-ACTIVATED PROTEIN KINASE KINASE KINASE 7-INTERACTING PROTEIN 2, UBIQUITIN, ZINC ION | | Authors: | Kulathu, Y, Akutsu, M, Bremm, A, Hofmann, K, Komander, D. | | Deposit date: | 2009-10-30 | | Release date: | 2009-11-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Two-Sided Ubiquitin Binding Explains Specificity of the Tab2 Nzf Domain
Nat.Struct.Mol.Biol., 16, 2009
|
|
2BKA
 
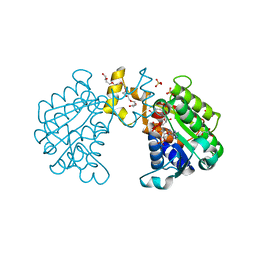 | | CC3(TIP30)Crystal Structure | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, GLYCEROL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | El Omari, K, Bird, L.E, Nichols, C.E, Ren, J, Stammers, D.K. | | Deposit date: | 2005-02-14 | | Release date: | 2005-02-21 | | Last modified: | 2016-12-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Cc3 (Tip30): Implications for its Role as a Tumor Suppressor
J.Biol.Chem., 280, 2005
|
|
2BJ4
 
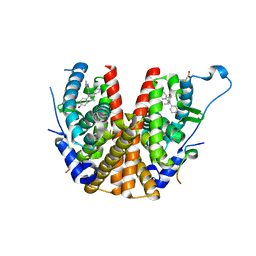 | | ESTROGEN RECEPTOR ALPHA LBD IN COMPLEX WITH A PHAGE-DISPLAY DERIVED PEPTIDE ANTAGONIST | | Descriptor: | 4-HYDROXYTAMOXIFEN, ESTROGEN RECEPTOR, PEPTIDE ANTAGONIST | | Authors: | Kong, E, Heldring, N, Gustafsson, J.A, Treuter, E, Hubbard, R.E, Pike, A.C.W. | | Deposit date: | 2005-01-28 | | Release date: | 2005-02-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Delineation of a Unique Protein-Protein Interaction Site on the Surface of the Estrogen Receptor
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2C2L
 
 | | Crystal structure of the CHIP U-box E3 ubiquitin ligase | | Descriptor: | CARBOXY TERMINUS OF HSP70-INTERACTING PROTEIN, HSP90, NICKEL (II) ION, ... | | Authors: | Zhang, M, Roe, S.M, Pearl, L.H. | | Deposit date: | 2005-09-29 | | Release date: | 2005-11-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Chaperoned Ubiquitylation-Crystal Structures of the Chip U Box E3 Ubiquitin Ligase and a Chip-Ubc13-Uev1A Complex
Mol.Cell, 20, 2005
|
|
2XPB
 
 | | DISCOVERY OF CELL-ACTIVE PHENYL-IMIDAZOLE PIN1 INHIBITORS BY STRUCTURE-GUIDED FRAGMENT EVOLUTION | | Descriptor: | 5-[BENZYL(METHYL)CARBAMOYL]-2-(3-CHLOROPHENYL)-1H-IMIDAZOLE-4-CARBOXYLIC ACID, DODECAETHYLENE GLYCOL, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE NIMA-INTERACTING 1 | | Authors: | Potter, A, Oldfield, V, Nunns, C, Fromont, C, Ray, S, Northfield, C.J, Bryant, C.J, Scrace, S.F, Robinson, D, Matossova, N, Baker, L, Dokurno, P, Surgenor, A.E, Davis, B.E, Richardson, C.M, Murray, J.B, Moore, J.D. | | Deposit date: | 2010-08-25 | | Release date: | 2011-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Cell-Active Phenyl-Imidazole Pin1 Inhibitors by Structure-Guided Fragment Evolution.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
7C36
 
 | | c-Myc DNA binding protein structure | | Descriptor: | RNA-binding motif, single-stranded-interacting protein 1 | | Authors: | Aggarwal, P, Bhavesh, N.S. | | Deposit date: | 2020-05-11 | | Release date: | 2021-05-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Hinge like domain motion facilitates human RBMS1 protein binding to proto-oncogene c-myc promoter.
Nucleic Acids Res., 49, 2021
|
|
4UIS
 
 | | The cryoEM structure of human gamma-Secretase complex | | Descriptor: | GAMMA-SECRETASE, LYSOZYME | | Authors: | Sun, L, Zhao, L, Yang, G, Yan, C, Zhou, R, Zhou, X, Xie, T, Zhao, Y, Wu, S, Li, X, Shi, Y. | | Deposit date: | 2015-04-03 | | Release date: | 2015-06-10 | | Last modified: | 2017-08-02 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural Basis of Human Gamma-Secretase Assembly.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7EA2
 
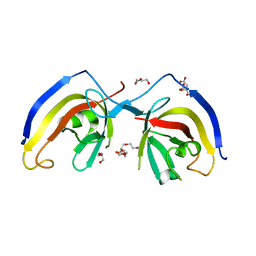 | | crystal structure of NAP1 FIR in complex with RB1CC1 Claw domain | | Descriptor: | 5-azacytidine-induced protein 2,RB1-inducible coiled-coil protein 1, CITRATE ANION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Fu, T, Pan, L. | | Deposit date: | 2021-03-06 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structural and biochemical advances on the recruitment of the autophagy-initiating ULK and TBK1 complexes by autophagy receptor NDP52.
Sci Adv, 7, 2021
|
|
7EAA
 
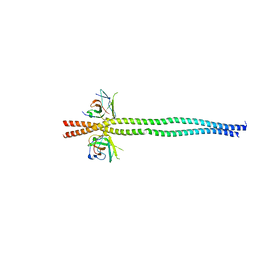 | | crystal structure of NDP52 SKICH domain in complex with RB1CC1 coiled-coil domain | | Descriptor: | Calcium-binding and coiled-coil domain-containing protein 2, RB1-inducible coiled-coil protein 1 | | Authors: | Fu, T, Pan, L. | | Deposit date: | 2021-03-06 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and biochemical advances on the recruitment of the autophagy-initiating ULK and TBK1 complexes by autophagy receptor NDP52.
Sci Adv, 7, 2021
|
|
2ZQV
 
 | |
