6DF2
 
 | |
7S0I
 
 | | CRYSTAL STRUCTURE OF N1 NEURAMINIDASE FROM A/Michigan/45/2015(H1N1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Neuraminidase, ... | | Authors: | Zhu, X, Wilson, I.A. | | Deposit date: | 2021-08-30 | | Release date: | 2021-12-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.892 Å) | | Cite: | A Novel Recombinant Influenza Virus Neuraminidase Vaccine Candidate Stabilized by a Measles Virus Phosphoprotein Tetramerization Domain Provides Robust Protection from Virus Challenge in the Mouse Model.
Mbio, 12, 2021
|
|
7SP5
 
 | | Crystal Structure of a Eukaryotic Phosphate Transporter | | Descriptor: | PHOSPHATE ION, Phosphate transporter, nonyl beta-D-glucopyranoside | | Authors: | Stroud, R.M, Pedersen, B.P, Kumar, H, Waight, A.B, Risenmay, A.J, Roe-Zurz, Z, Chau, B.H, Schlessinger, A, Bonomi, M, Harries, W, Sali, A, Johri, A.K, Finer-Moore, J. | | Deposit date: | 2021-11-02 | | Release date: | 2021-11-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of a eukaryotic phosphate transporter.
Nature, 496, 2013
|
|
7SHU
 
 | | IgE-Fc in complex with omalizumab variant C02 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoglobulin heavy constant epsilon, ... | | Authors: | Pennington, L.F, Jardetzky, T.J, Kleinboelting, S. | | Deposit date: | 2021-10-11 | | Release date: | 2021-12-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Directed evolution of and structural insights into antibody-mediated disruption of a stable receptor-ligand complex.
Nat Commun, 12, 2021
|
|
7SI0
 
 | | IgE-Fc in complex with 813 | | Descriptor: | 813 Variable fragment Heavy chain, 813 Variable fragment Light chain, IgE Fc, ... | | Authors: | Pennington, L.F, Jardetzky, T.J, Kleinboelting, S. | | Deposit date: | 2021-10-12 | | Release date: | 2021-12-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Directed evolution of and structural insights into antibody-mediated disruption of a stable receptor-ligand complex.
Nat Commun, 12, 2021
|
|
6DIX
 
 | | NFVFGT segment from Human Immunoglobulin Light-Chain Variable Domain, Residues 98-103, assembled as an amyloid fibril | | Descriptor: | NFVFGT Immunoglobulin Light-Chain Variable Domain | | Authors: | Brumshtein, B, Esswein, S.R, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2018-05-23 | | Release date: | 2018-10-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Identification of two principal amyloid-driving segments in variable domains of Ig light chains in systemic light-chain amyloidosis.
J. Biol. Chem., 293, 2018
|
|
7SHZ
 
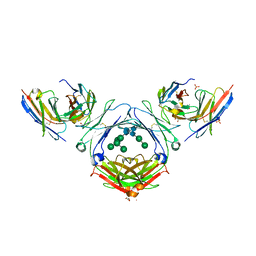 | | IgE-Fc in complex with HAE | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Pennington, L.F, Jardetzky, T.J, Kleinboelting, S. | | Deposit date: | 2021-10-11 | | Release date: | 2021-12-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Directed evolution of and structural insights into antibody-mediated disruption of a stable receptor-ligand complex.
Nat Commun, 12, 2021
|
|
6DMG
 
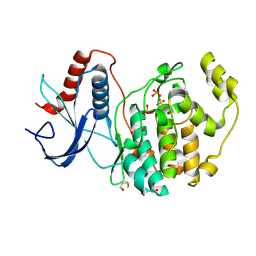 | | A multiconformer ligand model of EK6 bound to ERK2 | | Descriptor: | 1,2-ETHANEDIOL, Mitogen-activated protein kinase 1, SULFATE ION, ... | | Authors: | Hudson, B.M, van Zundert, G.C.P, Keedy, D.A, Fonseca, R, Heliou, A, Suresh, P, Borrelli, K, Day, T, Fraser, J.S, van den Bedem, H. | | Deposit date: | 2018-06-05 | | Release date: | 2018-12-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | qFit-ligand Reveals Widespread Conformational Heterogeneity of Drug-Like Molecules in X-Ray Electron Density Maps.
J. Med. Chem., 61, 2018
|
|
6DN3
 
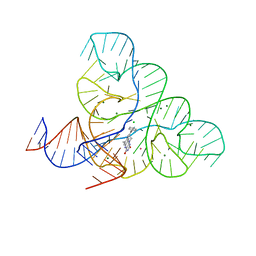 | | CRYSTAL STRUCTURE OF THE FMN RIBOSWITCH BOUND TO BRX1555 SPLIT RNA | | Descriptor: | 7,8-dimethyl-2,4-dioxo-10-(3-phenylpropyl)-1,2,3,4-tetrahydrobenzo[g]pteridin-10-ium, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Vicens, Q, Mondragon, E, Reyes, F.E, Berman, J, Kaur, H, Kells, K, Wickens, P, Wilson, J, Gadwood, R, Schostarez, H, Suto, R.K, Coish, P, Blount, K.F, Batey, R.T. | | Deposit date: | 2018-06-05 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-Activity Relationship of Flavin Analogues That Target the Flavin Mononucleotide Riboswitch.
ACS Chem. Biol., 13, 2018
|
|
9GSV
 
 | | Crystal structure of human lysosomal acid-alpha-glucosidase, GAA, in complex with iminosugar compound 4c | | Descriptor: | (2~{R},3~{R},4~{R},5~{S})-2-(hydroxymethyl)-2-(2-phenylethyl)piperidine-3,4,5-triol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sulzenbacher, G, Roig-Zamboni, V, Moracci, M, Parenti, G, Py, S. | | Deposit date: | 2024-09-16 | | Release date: | 2025-09-24 | | Last modified: | 2025-10-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | C -Branched Iminosugars as Selective Pharmacological Chaperones of Lysosomal alpha-Glucosidase for the Treatment of Pompe Disease.
J.Med.Chem., 68, 2025
|
|
7SPO
 
 | | Crystal structure of the SARS-CoV-2 receptor binding domain in complex with VNAR 3B4 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, ... | | Authors: | Shi, K, Aihara, H. | | Deposit date: | 2021-11-02 | | Release date: | 2022-01-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Mechanisms of SARS-CoV-2 neutralization by shark variable new antigen receptors elucidated through X-ray crystallography.
Nat Commun, 12, 2021
|
|
9GSW
 
 | | Crystal structure of human lysosomal acid-alpha-glucosidase, GAA, in complex with iminosugar compound 4d | | Descriptor: | (2~{R},3~{R},4~{R},5~{S})-2-(hydroxymethyl)-2-(2-trimethylsilylethyl)piperidine-3,4,5-triol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sulzenbacher, G, Roig-Zamboni, V, Moracci, M, Parenti, G, Py, S. | | Deposit date: | 2024-09-16 | | Release date: | 2025-09-24 | | Last modified: | 2025-10-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | C -Branched Iminosugars as Selective Pharmacological Chaperones of Lysosomal alpha-Glucosidase for the Treatment of Pompe Disease.
J.Med.Chem., 68, 2025
|
|
6DOK
 
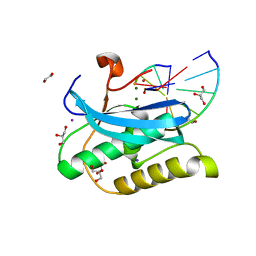 | |
9GTT
 
 | | Crystal structure of human lysosomal acid-alpha-glucosidase, GAA, in complex with iminosugar compound 4l | | Descriptor: | (2R,3R,4R,5S)-2-[2-(1-adamantyl)ethyl]-2-(hydroxymethyl)piperidine-3,4,5-triol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sulzenbacher, G, Roig-Zamboni, V, Moracci, M, Parenti, G, Py, S. | | Deposit date: | 2024-09-18 | | Release date: | 2025-10-01 | | Last modified: | 2025-10-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | C -Branched Iminosugars as Selective Pharmacological Chaperones of Lysosomal alpha-Glucosidase for the Treatment of Pompe Disease.
J.Med.Chem., 68, 2025
|
|
6DPC
 
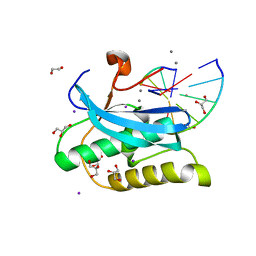 | |
7SLY
 
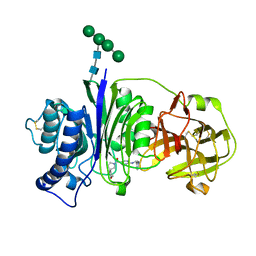 | | Vanin-1 complexed with Compound 27 | | Descriptor: | (8-oxa-2-azaspiro[4.5]decan-2-yl)(2-{[(1S)-1-(pyrazin-2-yl)ethyl]amino}pyrimidin-5-yl)methanone, 2-acetamido-2-deoxy-beta-D-glucopyranose, Pantetheinase, ... | | Authors: | Vajdos, F.F. | | Deposit date: | 2021-10-25 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Discovery of a Series of Pyrimidine Carboxamides as Inhibitors of Vanin-1.
J.Med.Chem., 65, 2022
|
|
8JL3
 
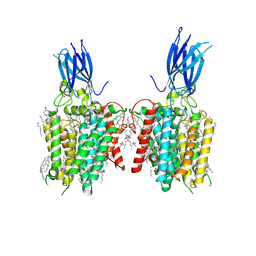 | | membrane proteins | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYL COENZYME *A, ... | | Authors: | Yu, J, Ge, J.P, Xu, R.S. | | Deposit date: | 2023-06-02 | | Release date: | 2024-05-22 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Structure and mechanism of lysosome transmembrane acetylation by HGSNAT.
Nat.Struct.Mol.Biol., 31, 2024
|
|
7SLX
 
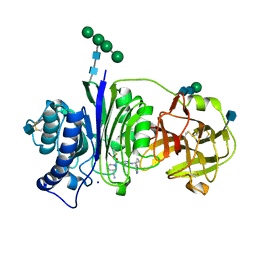 | | Vanin-1 complexed with Compound 11 | | Descriptor: | (3R)-1-(2-{[1-(pyrimidin-5-yl)cyclopropyl]amino}pyrimidine-5-carbonyl)piperidine-3-carbonitrile, 2-acetamido-2-deoxy-beta-D-glucopyranose, Pantetheinase, ... | | Authors: | Vajdos, F.F. | | Deposit date: | 2021-10-25 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery of a Series of Pyrimidine Carboxamides as Inhibitors of Vanin-1.
J.Med.Chem., 65, 2022
|
|
7SLV
 
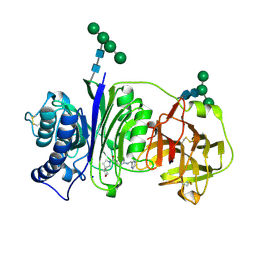 | | Vanin-1 complexed with Compound 3 | | Descriptor: | (8-oxa-2-azaspiro[4.5]decan-2-yl)(2-{[(pyrazin-2-yl)methyl]amino}pyrimidin-5-yl)methanone, 2-acetamido-2-deoxy-beta-D-glucopyranose, Pantetheinase, ... | | Authors: | Vajdos, F.F. | | Deposit date: | 2021-10-25 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Discovery of a Series of Pyrimidine Carboxamides as Inhibitors of Vanin-1.
J.Med.Chem., 65, 2022
|
|
6DQT
 
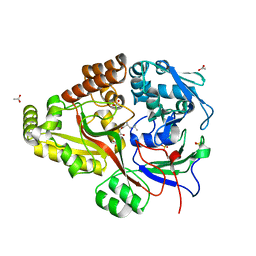 | | Crystal structure of Haemophilus influenzae OppA complex with LGG | | Descriptor: | ACETATE ION, LEU-GLY-GLY, Periplasmic oligopeptide-binding protein, ... | | Authors: | Tanaka, K.J, Pinkett, H.W. | | Deposit date: | 2018-06-11 | | Release date: | 2018-11-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Oligopeptide-binding protein from nontypeableHaemophilus influenzaehas ligand-specific sites to accommodate peptides and heme in the binding pocket.
J. Biol. Chem., 294, 2019
|
|
9GTC
 
 | | Crystal structure of human lysosomal acid-alpha-glucosidase, GAA, in complex with iminosugar compound 4g | | Descriptor: | (2~{R},3~{R},4~{R},5~{S})-2-(hydroxymethyl)-2-pentyl-piperidine-3,4,5-triol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sulzenbacher, G, Roig-Zamboni, V, Moracci, M, Parenti, G, Py, S. | | Deposit date: | 2024-09-17 | | Release date: | 2025-10-01 | | Last modified: | 2025-10-08 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | C -Branched Iminosugars as Selective Pharmacological Chaperones of Lysosomal alpha-Glucosidase for the Treatment of Pompe Disease.
J.Med.Chem., 68, 2025
|
|
9GTD
 
 | | Crystal structure of human lysosomal acid-alpha-glucosidase, GAA, in complex with iminosugar compound 4i | | Descriptor: | (2~{R},3~{R},4~{R},5~{S})-2-(2-cyclohexylethyl)-2-(hydroxymethyl)piperidine-3,4,5-triol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sulzenbacher, G, Roig-Zamboni, V, Moracci, M, Parenti, G, Py, S. | | Deposit date: | 2024-09-17 | | Release date: | 2025-10-01 | | Last modified: | 2025-10-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | C -Branched Iminosugars as Selective Pharmacological Chaperones of Lysosomal alpha-Glucosidase for the Treatment of Pompe Disease.
J.Med.Chem., 68, 2025
|
|
9GTN
 
 | | Crystal structure of human lysosomal acid-alpha-glucosidase, GAA, in complex with iminosugar compound 4k | | Descriptor: | (2~{R},3~{R},4~{R},5~{S})-2-(2-hydroxyethyl)-2-(hydroxymethyl)piperidine-3,4,5-triol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sulzenbacher, G, Roig-Zamboni, V, Moracci, M, Parenti, G, Py, S. | | Deposit date: | 2024-09-18 | | Release date: | 2025-10-01 | | Last modified: | 2025-10-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | C -Branched Iminosugars as Selective Pharmacological Chaperones of Lysosomal alpha-Glucosidase for the Treatment of Pompe Disease.
J.Med.Chem., 68, 2025
|
|
8JL4
 
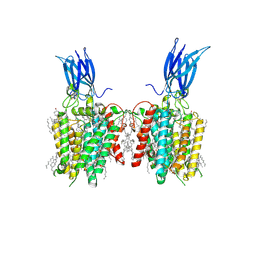 | | membrane proteins | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Yu, J, Ge, J.P, Xu, R.S. | | Deposit date: | 2023-06-02 | | Release date: | 2024-05-22 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | membrane proteins
To Be Published
|
|
9GTL
 
 | | Crystal structure of human lysosomal acid-alpha-glucosidase, GAA, in complex with iminosugar compound 4j | | Descriptor: | (2~{R},3~{R},4~{R},5~{S})-2-(hydroxymethyl)-2-(3-phenylpropyl)piperidine-3,4,5-triol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sulzenbacher, G, Roig-Zamboni, V, Moracci, M, Parenti, G, Py, S. | | Deposit date: | 2024-09-18 | | Release date: | 2025-10-01 | | Last modified: | 2025-10-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | C -Branched Iminosugars as Selective Pharmacological Chaperones of Lysosomal alpha-Glucosidase for the Treatment of Pompe Disease.
J.Med.Chem., 68, 2025
|
|
