1MSM
 
 | | The HIV protease (mutant Q7K L33I L63I) complexed with KNI-764 (an inhibitor) | | Descriptor: | (4R)-3-{(2S,3S)-2-hydroxy-3-[(3-hydroxy-2-methylbenzoyl)amino]-4-phenylbutanoyl}-5,5-dimethyl-N-(2-methylbenzyl)-1,3-thiazolidine-4-carboxamide, POL polyprotein | | Authors: | Vega, S, Kang, L.-W, Velazquez-Campoy, A, Kiso, Y, Amzel, L.M, Freire, E. | | Deposit date: | 2002-09-19 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A structural and thermodynamic escape mechanism from a drug resistant mutation of the HIV-1 protease.
Proteins, 55, 2004
|
|
7AOT
 
 | | The Fk1 domain of FKBP51 in complex with (2R,5S,12R)-12-cyclohexyl-2-[2-(3,4-dimethoxyphenyl)ethyl]-3,19-dioxa-10,13,16-triazatricyclo[18.3.1.0-5,10]tetracosa- 1(24),20,22-triene-4,11,14,17-tetrone | | Descriptor: | (2R,5S,12R)-12-cyclohexyl-2-[2-(3,4-dimethoxyphenyl)ethyl]-3,19-dioxa-10,13,16-triazatricyclo[18.3.1.0-5,10]tetracosa- 1(24),20,22-triene-4,11,14,17-tetrone, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Voll, A.M, Meyners, C, Heymann, T, Merz, S, Purder, P, Bracher, A, Hausch, F. | | Deposit date: | 2020-10-15 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Macrocyclic FKBP51 Ligands Define a Transient Binding Mode with Enhanced Selectivity.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7AWF
 
 | | The Fk1 domain of FKBP51 in complex with (2R,5S,12R)-12-cyclohexyl-2-[2-(3,4-dimethoxyphenyl)ethyl]-15,15,16-trimethyl-3,19-dioxa-10,13,16-triazatricyclo[18.3.1.0^5,^10]tetracosa-1(24),20,22-triene-4,11,14,17-tetrone | | Descriptor: | (2~{R},5~{S},12~{R})-12-cyclohexyl-2-[2-(3,4-dimethoxyphenyl)ethyl]-15,15,16-trimethyl-3,19-dioxa-10,13,16-triazatricyclo[18.3.1.0^{5,10}]tetracosa-1(24),20,22-triene-4,11,14,17-tetrone, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Voll, M.A, Meyners, C, Heymann, T, Merz, S, Purder, P, Bracher, A, Hausch, F. | | Deposit date: | 2020-11-07 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Macrocyclic FKBP51 Ligands Define a Transient Binding Mode with Enhanced Selectivity.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7AOU
 
 | | The Fk1 domain of FKBP51 in complex with (2'R,5'S,12'R)-12'-cyclohexyl-2'-[2-(3,4-dimethoxyphenyl)ethyl]-3',19'-dioxa-10',13',16'-triazaspiro[cyclopropane-1,15'- tricyclo[18.3.1.0-5,10]tetracosane]-1'(24'),20',22'-triene-4',11',14',17'-tetrone | | Descriptor: | (2'R,5'S,12'R)-12'-cyclohexyl-2'-[2-(3,4-dimethoxyphenyl)ethyl]-3',19'-dioxa-10',13',16'-triazaspiro[cyclopropane-1,15'- tricyclo[18.3.1.0-5,10]tetracosane]-1'(24'),20',22'-triene-4',11',14',17'-tetrone, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Voll, M.A, Meyners, C, Heymann, T, Merz, S, Purder, P, Bracher, A, Hausch, F. | | Deposit date: | 2020-10-15 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Macrocyclic FKBP51 Ligands Define a Transient Binding Mode with Enhanced Selectivity.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6V79
 
 | | Crystal structure of Danio rerio histone deacetylase 6 catalytic domain 2 (CD2) complexed with NF2376 | | Descriptor: | 1,2-ETHANEDIOL, 4-{[(2S)-3,3-dimethyl-2-(pyridin-3-yl)-2,3-dihydro-1H-indol-1-yl]methyl}-N-hydroxybenzamide, Hdac6 protein, ... | | Authors: | Osko, J.D, Christianson, D.W. | | Deposit date: | 2019-12-08 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.03951526 Å) | | Cite: | Harnessing the Role of HDAC6 in Idiopathic Pulmonary Fibrosis: Design, Synthesis, Structural Analysis, and Biological Evaluation of Potent Inhibitors.
J.Med.Chem., 64, 2021
|
|
4JJ0
 
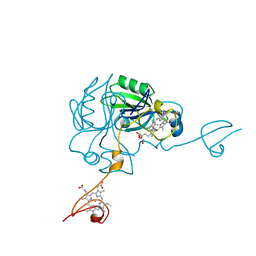 | | Crystal structure of MamP | | Descriptor: | GLYCEROL, HEME C, MamP | | Authors: | Siponen, M, Pignol, D, Arnoux, P. | | Deposit date: | 2013-03-07 | | Release date: | 2013-10-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insight into magnetochrome-mediated magnetite biomineralization.
Nature, 502, 2013
|
|
6Y1W
 
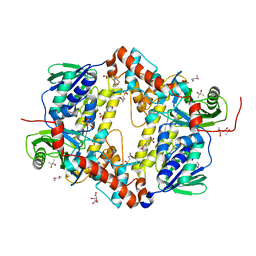 | | Xcc4156, a flavin-dependent halogenase from Xanthomonas campestris | | Descriptor: | (2S,3S)-butane-2,3-diol, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, L(+)-TARTARIC ACID, ... | | Authors: | Widmann, C, Ismail, M, Sewald, N, Niemann, H.H. | | Deposit date: | 2020-02-14 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of apo flavin-dependent halogenase Xcc4156 hints at a reason for cofactor-soaking difficulties.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
3QSB
 
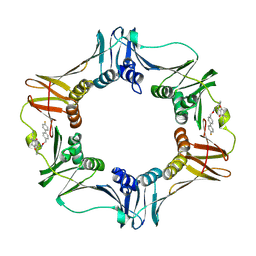 | | Structure of E. coli polIIIbeta with (Z)-5-(1-((4'-Fluorobiphenyl-4-yl)methoxyimino)butyl)-2,2-dimethyl-4,6-dioxocyclohexanecarbonitrile | | Descriptor: | (1R,5R)-5-{(1Z)-N-[(4'-fluorobiphenyl-4-yl)methoxy]butanimidoyl}-2,2-dimethyl-4,6-dioxocyclohexanecarbonitrile, DNA polymerase III subunit beta | | Authors: | Wijffels, G, Johnson, W.M, Oakley, A.J, Turner, K, Epa, V.C, Briscoe, S.J, Polley, M, Liepa, A.J, Hofmann, A, Buchardt, J, Christensen, C, Prosselkov, P, Dalrymple, B.P, Alewood, P.F, Jennings, P.A, Dixon, N.E, Winkler, D.A. | | Deposit date: | 2011-02-20 | | Release date: | 2011-06-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Binding inhibitors of the bacterial sliding clamp by design
J.Med.Chem., 54, 2011
|
|
1LOK
 
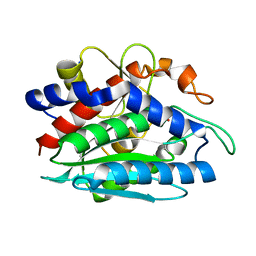 | | The 1.20 Angstrom Resolution Crystal Structure of the Aminopeptidase from Aeromonas proteolytica Complexed with Tris: A Tale of Buffer Inhibition | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Bacterial leucyl aminopeptidase, SODIUM ION, ... | | Authors: | Desmarais, W.T, Bienvenue, D.L, Bzymek, K.P, Holz, R.C, Petsko, G.A, Ringe, D. | | Deposit date: | 2002-05-06 | | Release date: | 2002-11-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The 1.20 Angstrom Resolution Crystal Structure of the Aminopeptidase from Aeromonas proteolytica Complexed with Tris A tale of Buffer Inhibition
Structure, 10, 2002
|
|
7N6V
 
 | |
4JY9
 
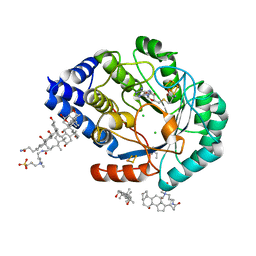 | | X-ray snapshots of possible intermediates in the time course of synthesis and degradation of protein-bound Fe4S4 clusters | | Descriptor: | CHAPSO, CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Nicolet, Y, Rohac, R, Martin, L, Fontecilla-Camps, J.C. | | Deposit date: | 2013-03-29 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray snapshots of possible intermediates in the time course of synthesis and degradation of protein-bound Fe4S4 clusters.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4JYF
 
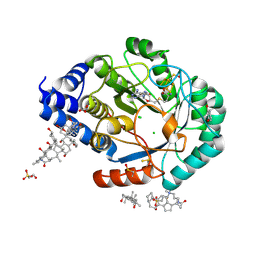 | | X-ray snapshots of possible intermediates in the time course of synthesis and degradation of protein-bound Fe4S4 clusters. | | Descriptor: | CARBONATE ION, CHAPSO, CHLORIDE ION, ... | | Authors: | Nicolet, Y, Rohac, R, Martin, L, Fontecilla-Camps, J.C. | | Deposit date: | 2013-03-29 | | Release date: | 2013-05-01 | | Last modified: | 2013-05-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | X-ray snapshots of possible intermediates in the time course of synthesis and degradation of protein-bound Fe4S4 clusters.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4JYE
 
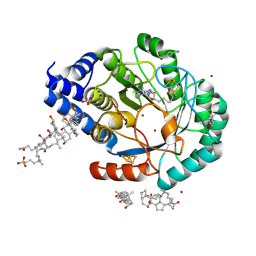 | | X-ray snapshots of possible intermediates in the time course of synthesis and degradation of protein-bound Fe4S4 clusters. | | Descriptor: | BROMIDE ION, Biotin synthetase, putative, ... | | Authors: | Nicolet, Y, Rohac, R, Martin, L, Fontecilla-Camps, J.C. | | Deposit date: | 2013-03-29 | | Release date: | 2013-05-01 | | Last modified: | 2013-05-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | X-ray snapshots of possible intermediates in the time course of synthesis and degradation of protein-bound Fe4S4 clusters.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4J5J
 
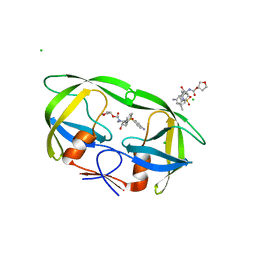 | |
3G65
 
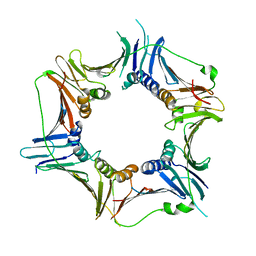 | | Crystal Structure of the Human Rad9-Rad1-Hus1 DNA Damage Checkpoint Complex | | Descriptor: | Cell cycle checkpoint control protein RAD9A, Cell cycle checkpoint protein RAD1, Checkpoint protein HUS1 | | Authors: | Dore, A.S, Kilkenny, M.L, Rzechorzek, N.J, Pearl, L.H. | | Deposit date: | 2009-02-06 | | Release date: | 2009-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the rad9-rad1-hus1 DNA damage checkpoint complex--implications for clamp loading and regulation.
Mol.Cell, 34, 2009
|
|
4JYD
 
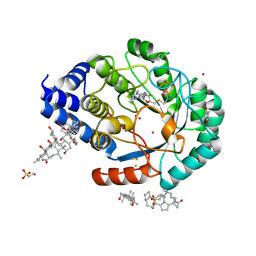 | | X-ray snapshots of possible intermediates in the time course of synthesis and degradation of protein-bound Fe4S4 clusters. | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, BROMIDE ION, CHAPSO, ... | | Authors: | Nicolet, Y, Rohac, R, Martin, L, Fontecilla-Camps, J.C. | | Deposit date: | 2013-03-29 | | Release date: | 2013-05-01 | | Last modified: | 2013-05-15 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | X-ray snapshots of possible intermediates in the time course of synthesis and degradation of protein-bound Fe4S4 clusters.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3GIT
 
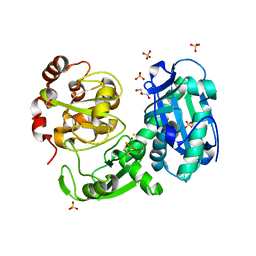 | | Crystal structure of a truncated acetyl-CoA synthase | | Descriptor: | Carbon monoxide dehydrogenase/acetyl-CoA synthase subunit alpha, GLYCEROL, HYDROSULFURIC ACID, ... | | Authors: | Volbeda, A, Darnault, C, Fontecilla-Camps, J.C. | | Deposit date: | 2009-03-06 | | Release date: | 2009-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Novel domain arrangement in the crystal structure of a truncated acetyl-CoA synthase from Moorella thermoacetica
Biochemistry, 48, 2009
|
|
8GS7
 
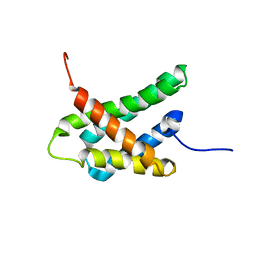 | | SOLUTION NMR STRUCTURE OF N-TERMINAL DOMAIN OF TRICONEPHILA CLAVIPES MAJOR AMPULLATE SPIDROIN 2 | | Descriptor: | Major ampullate spidroin 2 variant 3 | | Authors: | Oktaviani, N.A, Malay, A.D, Matsugami, A, Hayashi, F, Numata, K. | | Deposit date: | 2022-09-05 | | Release date: | 2023-03-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Unusual p K a Values Mediate the Self-Assembly of Spider Dragline Silk Proteins.
Biomacromolecules, 24, 2023
|
|
1SJX
 
 | | Three-Dimensional Structure of a Llama VHH Domain OE7 binding the cell wall protein Malf1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, immunoglobulin VH domain | | Authors: | Dolk, E, van der Vaart, M, Hulsik, D.L, Vriend, G, de Haard, H, Spinelli, S, Cambillau, C, Frenken, L, Verrips, T. | | Deposit date: | 2004-03-04 | | Release date: | 2005-03-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Isolation of llama antibody fragments for prevention of dandruff by phage display in shampoo.
Appl.Environ.Microbiol., 71, 2005
|
|
6R5F
 
 | | Crystal structure of RIP1 kinase in complex with DHP77 | | Descriptor: | Receptor-interacting serine/threonine-protein kinase 1, [(5~{S})-5-[3,5-bis(fluoranyl)phenyl]pyrazolidin-1-yl]-[1-(5-methyl-1,3,4-oxadiazol-2-yl)piperidin-4-yl]methanone | | Authors: | Thorpe, J.H, Campobasso, N, Harris, P.A. | | Deposit date: | 2019-03-25 | | Release date: | 2019-05-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Discovery and Lead-Optimization of 4,5-Dihydropyrazoles as Mono-Kinase Selective, Orally Bioavailable and Efficacious Inhibitors of Receptor Interacting Protein 1 (RIP1) Kinase.
J.Med.Chem., 62, 2019
|
|
4JJ3
 
 | |
4JY8
 
 | | X-ray snapshots of possible intermediates in the time course of synthesis and degradation of protein-bound Fe4S4 clusters | | Descriptor: | CHLORIDE ION, FEFE-HYDROGENASE MATURASE, HYDROSULFURIC ACID, ... | | Authors: | Nicolet, Y, Rohac, R, Martin, L, Fontecilla-Camps, J.C. | | Deposit date: | 2013-03-29 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | X-ray snapshots of possible intermediates in the time course of synthesis and degradation of protein-bound Fe4S4 clusters.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1P0Q
 
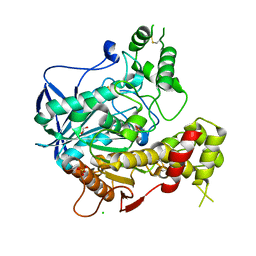 | | Crystal structure of soman-aged human butyryl cholinesterase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Nicolet, Y, Lockridge, O, Masson, P, Fontecilla-Camps, J.C, Nachon, F. | | Deposit date: | 2003-04-10 | | Release date: | 2003-08-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Crystal structure of human butyrylcholinesterase and of its complexes with substrate and products.
J.Biol.Chem., 278, 2003
|
|
6VZK
 
 | |
6TL2
 
 | |
