1KG4
 
 | | Crystal structure of the K142A mutant of E. coli MutY (core fragment) | | Descriptor: | A/G-specific adenine glycosylase, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Gilboa, R, Kilshtein, A, Zharkov, D.O, Kycia, J.H, Gerchman, S.E, Grollman, A.P, Shoham, G. | | Deposit date: | 2001-11-26 | | Release date: | 2002-11-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Analysis of the E.coli MutY DNA glycosylase structure and function by site-directed mutagenesis
To be Published
|
|
7AK0
 
 | | Human MALT1(329-729) in complex with a chromane urea containing inhibitor | | Descriptor: | 1-[4-[4-(aminomethyl)pyrazol-1-yl]-3-chloranyl-phenyl]-3-[(3~{R})-6-bromanyl-3,4-dihydro-2~{H}-chromen-3-yl]urea, Mucosa-associated lymphoid tissue lymphoma translocation protein 1 | | Authors: | Renatus, M. | | Deposit date: | 2020-09-29 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.316 Å) | | Cite: | Discovery of Potent, Highly Selective, and In Vivo Efficacious, Allosteric MALT1 Inhibitors by Iterative Scaffold Morphing.
J.Med.Chem., 63, 2020
|
|
1KTS
 
 | | Thrombin Inhibitor Complex | | Descriptor: | 3-({2-[(4-CARBAMIMIDOYL-PHENYLAMINO)-METHYL]-3-METHYL-3H-BENZOIMIDAZOLE-5-CARBONYL}-PYRIDIN-2-YL-AMINO)-PROPIONIC ACID ETHYL ESTER, hirudin IIB, thrombin | | Authors: | Nar, H. | | Deposit date: | 2002-01-17 | | Release date: | 2002-02-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based design of novel potent nonpeptide thrombin inhibitors.
J.Med.Chem., 45, 2002
|
|
1KU0
 
 | | Structure of the Bacillus stearothermophilus L1 lipase | | Descriptor: | CALCIUM ION, L1 lipase, ZINC ION | | Authors: | Jeong, S.-T, Kim, H.-K, Kim, S.-J, Chi, S.-W, Pan, J.-G, Oh, T.-K, Ryu, S.-E. | | Deposit date: | 2002-01-18 | | Release date: | 2002-08-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Novel zinc-binding center and a temperature switch in the Bacillus stearothermophilus L1 lipase.
J.Biol.Chem., 277, 2002
|
|
7AFS
 
 | | The structure of Artemis variant D37A | | Descriptor: | 1,2-ETHANEDIOL, NICKEL (II) ION, Protein artemis, ... | | Authors: | Yosaatmadja, Y, Goubin, S, Newman, J.A, Mukhopadhyay, S.M.M, Dannerfjord, A.A, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-20 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and mechanistic insights into the Artemis endonuclease and strategies for its inhibition.
Nucleic Acids Res., 49, 2021
|
|
9CUZ
 
 | | Bufavirus 1 complexed with 6SLN | | Descriptor: | N-acetyl-alpha-neuraminic acid, VP1 | | Authors: | Gulkis, M.C, McKenna, R, Bennett, A.D. | | Deposit date: | 2024-07-27 | | Release date: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (2.16 Å) | | Cite: | Structural Characterization of Human Bufavirus 1: Receptor Binding and Endosomal pH-Induced Changes.
Viruses, 16, 2024
|
|
1KUT
 
 | | Structural Genomics, Protein TM1243, (SAICAR synthetase) | | Descriptor: | Phosphoribosylaminoimidazole-succinocarboxamide synthase | | Authors: | Zhang, R, Skarina, T, Beasley, S, Edwards, A, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-01-22 | | Release date: | 2002-08-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of SAICAR synthase from Thermotoga maritima at 2.2 angstroms reveals an unusual covalent dimer.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
1KVL
 
 | | X-ray Crystal Structure of AmpC S64G Mutant beta-Lactamase in Complex with Substrate and Product Forms of Cephalothin | | Descriptor: | 2-[CARBOXY-(2-THIOPHEN-2-YL-ACETYLAMINO)-METHYL]-5-METHYL-3,6-DIHYDRO-2H-[1,3]THIAZINE-4-CARBOXYLIC ACID, 2-[CARBOXY-(2-THIOPHEN-2-YL-ACETYLAMINO)-METHYL]-5-METHYLENE-5,6-DIHYDRO-2H-[1,3]THIAZINE-4-CARBOXYLIC ACID, Beta-lactamase, ... | | Authors: | Beadle, B.M, Trehan, I, Focia, P.J, Shoichet, B.K. | | Deposit date: | 2002-01-27 | | Release date: | 2002-03-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structural milestones in the reaction pathway of an amide hydrolase: substrate, acyl, and product complexes of cephalothin with AmpC beta-lactamase.
Structure, 10, 2002
|
|
1KGB
 
 | | structure of ground-state bacteriorhodopsin | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, RETINAL, bacteriorhodopsin | | Authors: | Facciotti, M.T, Rouhani, S, Burkard, F.T, Betancourt, F.M, Downing, K.H, Rose, R.B, McDermott, G, Glaeser, R.M. | | Deposit date: | 2001-11-26 | | Release date: | 2001-12-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of an early intermediate in the M-state phase of the bacteriorhodopsin photocycle.
Biophys.J., 81, 2001
|
|
1KGQ
 
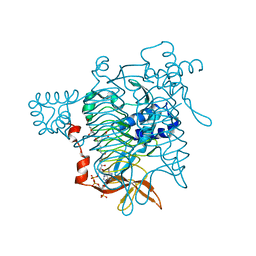 | | Crystal Structure of Tetrahydrodipicolinate N-Succinyltransferase in Complex with L-2-aminopimelate and Succinamide-CoA | | Descriptor: | (2S)-2-aminoheptanedioic acid, 2,3,4,5-TETRAHYDROPYRIDINE-2-CARBOXYLATE N-SUCCINYLTRANSFERASE, SUCCINAMIDE-COA | | Authors: | Beaman, T.W, Vogel, K.W, Drueckhammer, D.G, Blanchard, J.S, Roderick, S.L. | | Deposit date: | 2001-11-28 | | Release date: | 2002-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Acyl group specificity at the active site of tetrahydridipicolinate N-succinyltransferase.
Protein Sci., 11, 2002
|
|
1KDX
 
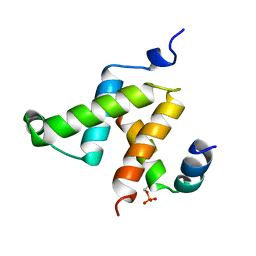 | | KIX DOMAIN OF MOUSE CBP (CREB BINDING PROTEIN) IN COMPLEX WITH PHOSPHORYLATED KINASE INDUCIBLE DOMAIN (PKID) OF RAT CREB (CYCLIC AMP RESPONSE ELEMENT BINDING PROTEIN), NMR 17 STRUCTURES | | Descriptor: | CBP, CREB | | Authors: | Radhakrishnan, I, Perez-Alvarado, G.C, Dyson, H.J, Wright, P.E. | | Deposit date: | 1997-09-16 | | Release date: | 1998-11-25 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the KIX domain of CBP bound to the transactivation domain of CREB: a model for activator:coactivator interactions.
Cell(Cambridge,Mass.), 91, 1997
|
|
1KDH
 
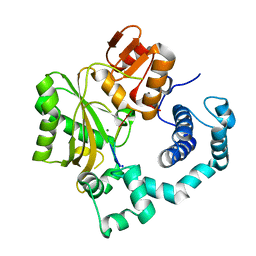 | | Binary Complex of Murine Terminal Deoxynucleotidyl Transferase with a Primer Single Stranded DNA | | Descriptor: | 5'-D(P*(BRU)P*(BRU)P*(BRU)P*(BRU))-3', MAGNESIUM ION, SODIUM ION, ... | | Authors: | Delarue, M, Boule, J.B, Lescar, J, Expert-Bezancon, N, Jourdan, N, Sukumar, N, Rougeon, F, Papanicolaou, C. | | Deposit date: | 2001-11-13 | | Release date: | 2002-05-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of a template-independent DNA polymerase: murine terminal deoxynucleotidyltransferase.
EMBO J., 21, 2002
|
|
7AN4
 
 | |
1KH2
 
 | |
1KHQ
 
 | | ORTHORHOMBIC FORM OF PAPAIN/ZLFG-DAM COVALENT COMPLEX | | Descriptor: | Papain, peptidic inhibitor | | Authors: | Janowski, R, Kozak, M, Jankowska, E, Grzonka, Z, Jaskolski, M. | | Deposit date: | 2001-11-30 | | Release date: | 2003-09-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Two polymorphs of a covalent complex between papain and a diazomethylketone inhibitor
J.Pept.Res., 64, 2004
|
|
7AQ8
 
 | | Pseudomonas stutzeri nitrous oxide reductase mutant, H583Y/D576A | | Descriptor: | (MU-4-SULFIDO)-TETRA-NUCLEAR COPPER ION, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Zhang, L, Bill, E, Kroneck, P.M.H, Einsle, O. | | Deposit date: | 2020-10-20 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.791 Å) | | Cite: | Histidine-Gated Proton-Coupled Electron Transfer to the Cu A Site of Nitrous Oxide Reductase.
J.Am.Chem.Soc., 143, 2021
|
|
1KDS
 
 | |
7ANJ
 
 | |
6VRR
 
 | |
1KE9
 
 | | CYCLIN-DEPENDENT KINASE 2 (CDK2) COMPLEXED WITH 3-{[4-({[AMINO(IMINO)METHYL]AMINOSULFONYL)ANILINO]METHYLENE}-2-OXO-2,3-DIHYDRO-1H-INDOLE | | Descriptor: | 3-{[4-([AMINO(IMINO)METHYL]AMINOSULFONYL)ANILINO]METHYLENE}-2-OXO-2,3-DIHYDRO-1H-INDOLE, Cell division protein kinase 2 | | Authors: | Bramson, H.N, Corona, J, Davis, S.T, Dickerson, S.H, Edelstein, M, Frye, S.V, Gampe, R.T, Hassell, A.M, Shewchuk, L.M, Kuyper, L.F. | | Deposit date: | 2001-11-14 | | Release date: | 2002-05-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Oxindole-based inhibitors of cyclin-dependent kinase 2 (CDK2): design, synthesis, enzymatic activities, and X-ray crystallographic analysis.
J.Med.Chem., 44, 2001
|
|
1KHX
 
 | | Crystal structure of a phosphorylated Smad2 | | Descriptor: | Smad2 | | Authors: | Wu, J.-W, Hu, M, Chai, J, Seoane, J, Huse, M, Kyin, S, Muir, T.W, Fairman, R, Massague, J, Shi, Y. | | Deposit date: | 2001-12-01 | | Release date: | 2002-02-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a phosphorylated Smad2. Recognition of phosphoserine by the MH2 domain and insights on Smad function in TGF-beta signaling.
Mol.Cell, 8, 2001
|
|
7AON
 
 | | Crystal structure of CI2 double mutant L49I,I57V | | Descriptor: | GLYCEROL, SULFATE ION, Subtilisin-chymotrypsin inhibitor-2A | | Authors: | Olsen, J.G, Teilum, K, Hamborg, L, Roche, J.V. | | Deposit date: | 2020-10-14 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Synergistic stabilization of a double mutant in chymotrypsin inhibitor 2 from a library screen in E. coli.
Commun Biol, 4, 2021
|
|
1KI7
 
 | | CRYSTAL STRUCTURE OF THYMIDINE KINASE FROM HERPES SIMPLEX VIRUS TYPE I COMPLEXED WITH 5-IODODEOXYURIDINE | | Descriptor: | 5-IODODEOXYURIDINE, SULFATE ION, THYMIDINE KINASE | | Authors: | Champness, J.N, Bennett, M.S, Wien, F, Visse, R, Summers, W.C, Sanderson, M.R. | | Deposit date: | 1998-05-15 | | Release date: | 1998-12-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Exploring the active site of herpes simplex virus type-1 thymidine kinase by X-ray crystallography of complexes with aciclovir and other ligands.
Proteins, 32, 1998
|
|
1KEJ
 
 | | Crystal Structure of Murine Terminal Deoxynucleotidyl Transferase complexed with ddATP | | Descriptor: | 2',3'-DIDEOXYADENOSINE-5'-TRIPHOSPHATE, COBALT (II) ION, SODIUM ION, ... | | Authors: | Delarue, M, Boule, J.B, Lescar, J, Expert-Bezancon, N, Jourdan, N, Sukumar, N, Rougeon, F, Papanicolaou, C. | | Deposit date: | 2001-11-16 | | Release date: | 2002-05-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of a template-independent DNA polymerase: murine terminal deoxynucleotidyltransferase.
EMBO J., 21, 2002
|
|
1KEP
 
 | | The crystal structure of dTDP-D-glucose 4,6-dehydratase (RmlB) from Streptococcus suis with dTDP-xylose bound | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, THYMIDINE-5'-DIPHOSPHO-BETA-D-XYLOSE, ... | | Authors: | Allard, S.T.M, Beis, K, Giraud, M.-F, Hegeman, A.D, Gross, J.W, Whitfield, C, Graninger, M, Messner, P, Allen, A.G, Naismith, J.H. | | Deposit date: | 2001-11-16 | | Release date: | 2002-01-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Toward a structural understanding of the dehydratase mechanism.
Structure, 10, 2002
|
|
