1C3U
 
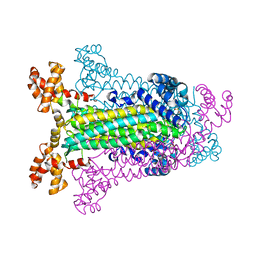 | | T. MARITIMA ADENYLOSUCCINATE LYASE | | Descriptor: | ADENYLOSUCCINATE LYASE, SULFATE ION | | Authors: | Toth, E.A, Yeates, T.O. | | Deposit date: | 1999-07-28 | | Release date: | 2000-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of adenylosuccinate lyase, an enzyme with dual activity in the de novo purine biosynthetic pathway.
Structure Fold.Des., 8, 2000
|
|
6ARY
 
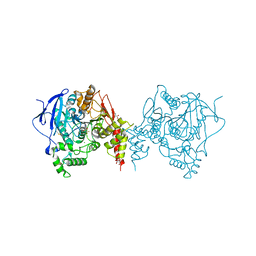 | | Crystal structure of an insecticide-resistant acetylcholinesterase mutant from the malaria vector Anopheles gambiae in complex with a difluoromethyl ketone inhibitor | | Descriptor: | (1S)-2,2-difluoro-1-[1-(pentan-3-yl)-1H-pyrazol-4-yl]ethan-1-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, ... | | Authors: | Cheung, J, Mahmood, A, Kalathur, R, Lixuan, L, Carlier, P.R. | | Deposit date: | 2017-08-23 | | Release date: | 2018-01-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.257 Å) | | Cite: | Structure of the G119S Mutant Acetylcholinesterase of the Malaria Vector Anopheles gambiae Reveals Basis of Insecticide Resistance.
Structure, 26, 2018
|
|
6ARX
 
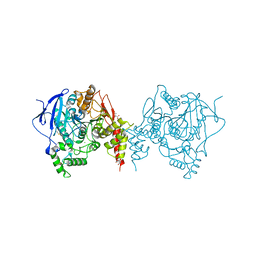 | | Crystal structure of an insecticide-resistant acetylcholinesterase mutant from the malaria vector Anopheles gambiae in the ligand-free state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, CHLORIDE ION, ... | | Authors: | Cheung, J, Mahmood, A, Kalathur, R, Lixuan, L, Carlier, P.R. | | Deposit date: | 2017-08-23 | | Release date: | 2018-01-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Structure of the G119S Mutant Acetylcholinesterase of the Malaria Vector Anopheles gambiae Reveals Basis of Insecticide Resistance.
Structure, 26, 2018
|
|
6RGZ
 
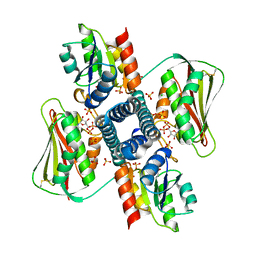 | | Revisiting pH-gated conformational switch. Complex HK853-RR468 pH 6.5 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Mideros-Mora, C, Casino, P, Marina, A. | | Deposit date: | 2019-04-18 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Revisiting the pH-gated conformational switch on the activities of HisKA-family histidine kinases.
Nat Commun, 11, 2020
|
|
6RGY
 
 | | Revisiting pH-gated conformational switch. Complex HK853-RR468 pH 7.5 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CITRIC ACID, MAGNESIUM ION, ... | | Authors: | Mideros-Mora, C, Casino, P, Marina, A. | | Deposit date: | 2019-04-18 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Revisiting the pH-gated conformational switch on the activities of HisKA-family histidine kinases.
Nat Commun, 11, 2020
|
|
4REY
 
 | | Crystal Structure of the GRASP65-GM130 C-terminal peptide complex | | Descriptor: | Golgi reassembly-stacking protein 1, Golgin subfamily A member 2, SULFATE ION | | Authors: | Shi, N, Hu, F, Li, B. | | Deposit date: | 2014-09-24 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural Basis for the Interaction between the Golgi Reassembly-stacking Protein GRASP65 and the Golgi Matrix Protein GM130.
J.Biol.Chem., 290, 2015
|
|
3ZR4
 
 | | STRUCTURAL EVIDENCE FOR AMMONIA TUNNELING ACROSS THE (BETA-ALPHA)8 BARREL OF THE IMIDAZOLE GLYCEROL PHOSPHATE SYNTHASE BIENZYME COMPLEX | | Descriptor: | GLUTAMINE, GLYCEROL, IMIDAZOLE GLYCEROL PHOSPHATE SYNTHASE SUBUNIT HISF, ... | | Authors: | Vega, M.C, Kuper, J, Haeger, M.C, Mohrlueder, J, Marquardt, S, Sterner, R, Wilmanns, M. | | Deposit date: | 2011-06-13 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Catalysis Uncoupling in a Glutamine Amidotransferase Bienzyme by Unblocking the Glutaminase Active Site.
Chem.Biol., 19, 2012
|
|
6IW8
 
 | | Crystal structure of Importin-alpha and wild-type GM130 | | Descriptor: | Importin subunit alpha-1, Peptide from Golgin subfamily A member 2 | | Authors: | Chang, C.-C, Chen, C.-J, Pien, Y.-C, Tsai, S.-Y, Hsia, K.-C. | | Deposit date: | 2018-12-04 | | Release date: | 2019-10-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Ran pathway-independent regulation of mitotic Golgi disassembly by Importin-alpha.
Nat Commun, 10, 2019
|
|
7KOE
 
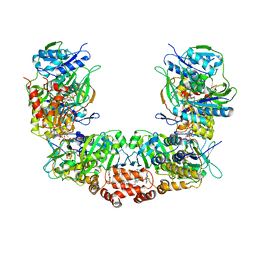 | | Electron bifurcating flavoprotein Fix/EtfABCX | | Descriptor: | Electron transfer flavoprotein, alpha subunit, beta subunit, ... | | Authors: | Feng, X, Li, H. | | Deposit date: | 2020-11-08 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryoelectron microscopy structure and mechanism of the membrane-associated electron-bifurcating flavoprotein Fix/EtfABCX.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6IWA
 
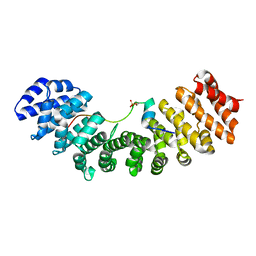 | | Crystal structure of Importin-alpha and phosphoserine GM130 | | Descriptor: | Importin subunit alpha-1, PHOSPHOSERINE GM130 | | Authors: | Chang, C.-C, Chen, C.-J, Pien, Y.-C, Tsai, S.-Y, Hsia, K.-C. | | Deposit date: | 2018-12-05 | | Release date: | 2019-10-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Ran pathway-independent regulation of mitotic Golgi disassembly by Importin-alpha.
Nat Commun, 10, 2019
|
|
6K06
 
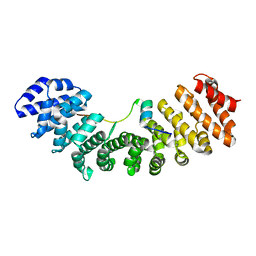 | | Crystal structure of Importin-alpha and phosphomimetic GM130 | | Descriptor: | Importin subunit alpha-1, Peptide from Golgin subfamily A member 2 | | Authors: | Chang, C.-C, Chen, C.-J, Pien, Y.-C, Tsai, S.-Y, Hsia, K.-C. | | Deposit date: | 2019-05-05 | | Release date: | 2019-10-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Ran pathway-independent regulation of mitotic Golgi disassembly by Importin-alpha.
Nat Commun, 10, 2019
|
|
5X61
 
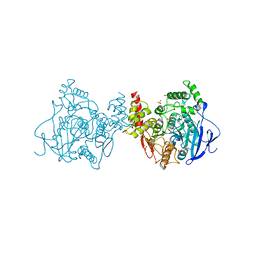 | | Crystal structure of Acetylcholinesterase Catalytic Subunit of the Malaria Vector Anopheles Gambiae, 3.4 A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, AMMONIUM ION, ... | | Authors: | Han, Q, Robinson, H, Ding, H, Wong, D.M, Lam, P.C.H, Totrov, M.M, Carlier, P.R, Li, J. | | Deposit date: | 2017-02-20 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal structure of acetylcholinesterase catalytic subunits of the malaria vector Anopheles gambiae
Insect Sci., 2017
|
|
5YDJ
 
 | | Crystal structure of anopheles gambiae acetylcholinesterase in complex with PMSF | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, ... | | Authors: | Han, Q, Guan, H, Ding, H, Liao, C, Robinson, H, Li, J. | | Deposit date: | 2017-09-13 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Crystal structures of acetylcholinesterase of the malaria vector Anopheles gambiae reveal a polymerization interface, ligand binding residues and post translational modifications
To Be Published
|
|
5YDI
 
 | | Crystal structure of acetylcholinesterase catalytic subunits of the malaria vector anopheles gambiae, new crystal packing | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, GLYCEROL, ... | | Authors: | Han, Q, Guan, H, Ding, H, Liao, C, Robinson, H, Li, J. | | Deposit date: | 2017-09-13 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Crystal structures of acetylcholinesterase of the malaria vector Anopheles gambiae reveal a polymerization interface, ligand binding residues and post translational modifications
To Be Published
|
|
5YDH
 
 | | Crystal structure of acetylcholinesterase catalytic subunits of the malaria vector anopheles gambiae, 3.2 A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Acetylcholinesterase, ... | | Authors: | Han, Q, Guan, H, Robinson, H, Ding, H, Liao, C, Li, J. | | Deposit date: | 2017-09-13 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Crystal structures of acetylcholinesterase of the malaria vector Anopheles gambiae reveal a polymerization interface, ligand binding residues and post translational modifications
To Be Published
|
|
2NRR
 
 | |
2NRX
 
 | | Crystal structure of the C-terminal half of UvrC, in the presence of sulfate molecules | | Descriptor: | GLYCEROL, SULFATE ION, UvrABC system protein C | | Authors: | Karakas, E, Truglio, J.J, Kisker, C. | | Deposit date: | 2006-11-02 | | Release date: | 2007-02-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the C-terminal half of UvrC reveals an RNase H endonuclease domain with an Argonaute-like catalytic triad.
Embo J., 26, 2007
|
|
2NRZ
 
 | |
2NRT
 
 | |
2NZ3
 
 | |
2NY9
 
 | |
2NRW
 
 | |
2NY8
 
 | |
7AC8
 
 | |
2NRV
 
 | |
