6RG7
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complexe with an inhibitor | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-[[3-(6-azanyl-9~{H}-purin-8-yl)prop-2-ynyl-methyl-amino]methyl]oxolane-3,4-diol, CITRIC ACID, NAD kinase 1 | | Authors: | Gelin, M, Labesse, G. | | Deposit date: | 2019-04-16 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | From Substrate to Fragments to Inhibitor ActiveIn VivoagainstStaphylococcus aureus.
Acs Infect Dis., 6, 2020
|
|
6PIW
 
 | |
6RGB
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complexe with an inhibitor | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-[[3-[6-azanyl-9-[(2~{R},3~{R},4~{S},5~{R})-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]purin-8-yl]prop-2-ynylamino]methyl]oxolane-3,4-diol, CITRIC ACID, NAD kinase 1 | | Authors: | Gelin, M, Labesse, G. | | Deposit date: | 2019-04-16 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | From Substrate to Fragments to Inhibitor ActiveIn VivoagainstStaphylococcus aureus.
Acs Infect Dis., 6, 2020
|
|
6DAY
 
 | | Xanthomonas albilineans Dihydropteroate synthase with 4-aminobenzoic acid at 1.65 A | | Descriptor: | 4-AMINOBENZOIC ACID, Dihydropteroate synthase, GLYCEROL, ... | | Authors: | Oliveira, A.A, Guido, R.V.C, Lima, G.M.A, Bueno, R.V, Maluf, F.V. | | Deposit date: | 2018-05-02 | | Release date: | 2018-06-06 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Xanthomonas albilineans Dihydropteroate synthase with 4-aminobenzoic acid
To Be Published
|
|
6P14
 
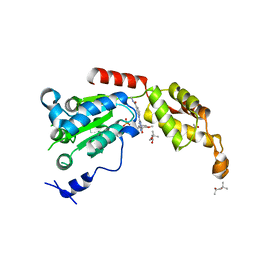 | | Structure of spastin AAA domain (T692A mutant) in complex with a diaminotriazole-based inhibitor (crystal form B) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-{[5-amino-1-(2-fluoro-6-methoxybenzene-1-carbonyl)-1H-1,2,4-triazol-3-yl]amino}-N-methylbenzamide, ACETATE ION, ... | | Authors: | Pisa, R, Cupido, T, Kapoor, T.M. | | Deposit date: | 2019-05-17 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.93001127 Å) | | Cite: | Analyzing Resistance to Design Selective Chemical Inhibitors for AAA Proteins.
Cell Chem Biol, 26, 2019
|
|
6FZ4
 
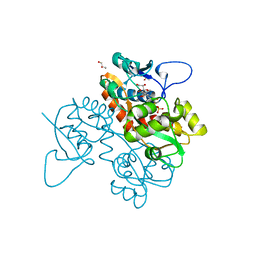 | | Structure of GluK1 ligand-binding domain in complex with N-(7-fluoro-2,3-dioxo-6-(trifluoromethyl)-3,4-dihydroquinoxalin-1(2H)-yl)-2-hydroxybenzamide at 1.85 A resolution | | Descriptor: | CHLORIDE ION, GLYCEROL, Glutamate receptor ionotropic, ... | | Authors: | Kastrup, J.S, Frydenvang, K, Mollerud, S. | | Deposit date: | 2018-03-14 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | N1-Substituted Quinoxaline-2,3-diones as Kainate Receptor Antagonists: X-ray Crystallography, Structure-Affinity Relationships, and in Vitro Pharmacology.
Acs Chem Neurosci, 10, 2019
|
|
9F0Q
 
 | | VIM-2 in complex with GKV53 (5d) - dynamically chiral phosphonic acid-type metallo-beta-lactamase inhibitors | | Descriptor: | FORMIC ACID, Metallo-beta-lactamase type 2, ZINC ION, ... | | Authors: | Bartels, K, Prester, A, Bosman, R, Gulyas, K.V, Erdelyi, M, Schulz, E.C. | | Deposit date: | 2024-04-17 | | Release date: | 2025-04-30 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Dynamically chiral phosphonic acid-type metallo-beta-lactamase inhibitors.
Commun Chem, 8, 2025
|
|
8DLJ
 
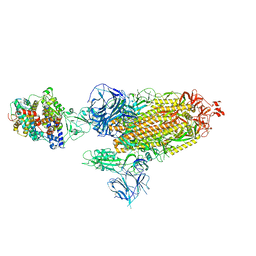 | | Cryo-EM structure of SARS-CoV-2 Alpha (B.1.1.7) spike protein in complex with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2022-08-31 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | SARS-CoV-2 variants of concern: spike protein mutational analysis and epitope for broad neutralization.
Nat Commun, 13, 2022
|
|
5ICE
 
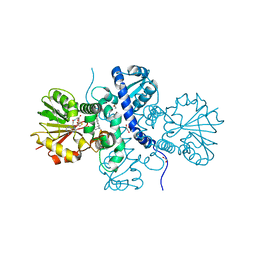 | | Crystal structure of (S)-norcoclaurine 6-O-methyltransferase with S-adenosyl-L-homocysteine and norlaudanosoline | | Descriptor: | (1S)-1-(3,4-dihydroxybenzyl)-1,2,3,4-tetrahydroisoquinoline-6,7-diol, (S)-norcoclaurine 6-O-methyltransferase, 1,2-ETHANEDIOL, ... | | Authors: | Robin, A.Y, Graindorge, M, Giustini, C, Dumas, R, Matringe, M. | | Deposit date: | 2016-02-23 | | Release date: | 2016-06-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of norcoclaurine-6-O-methyltransferase, a key rate-limiting step in the synthesis of benzylisoquinoline alkaloids.
Plant J., 87, 2016
|
|
9F0I
 
 | | Human Cyclophilin D in complex with fragment | | Descriptor: | Peptidyl-prolyl cis-trans isomerase F, mitochondrial, ~{N}-(2-ethyl-1,2,3,4-tetrazol-5-yl)thiophene-2-carboxamide | | Authors: | Silva, D.O, Graedler, U, Bandeiras, T.M. | | Deposit date: | 2024-04-16 | | Release date: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Human Cyclophilin D in complex with fragment
To Be Published
|
|
8DLU
 
 | | Cryo-EM structure of SARS-CoV-2 Epsilon (B.1.429) spike protein in complex with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2022-08-31 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | SARS-CoV-2 variants of concern: spike protein mutational analysis and epitope for broad neutralization.
Nat Commun, 13, 2022
|
|
6IRE
 
 | | Complex structure of INAD PDZ45 and NORPA CC-PBM | | Descriptor: | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase, Inactivation-no-after-potential D protein | | Authors: | Ye, F, Li, J, Deng, X, Liu, W, Zhang, M. | | Deposit date: | 2018-11-12 | | Release date: | 2019-01-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | An unexpected INAD PDZ tandem-mediated plc beta binding in Drosophila photo receptors.
Elife, 7, 2018
|
|
8ZBZ
 
 | | SARS-CoV-2 Omicron BA.2 spike trimer (6P) in complex with 3 D1F6 Fabs (1 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of D1F6 Fab, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, Q, He, J, Xiong, X. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-15 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (4.71 Å) | | Cite: | An unconventional VH1-2 antibody tolerates escape mutations and shows an antigenic hotspot on SARS-CoV-2 spike.
Cell Rep, 43, 2024
|
|
9C3Y
 
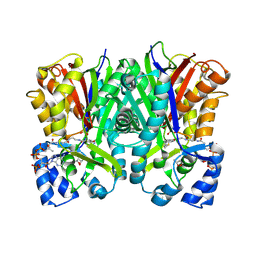 | | Crystal structure of biphenyl synthase from Malus domestica complexed with tetraketide-CoA mimetic | | Descriptor: | (3R)-butane-1,3-diol, BIS3 biphenyl synthase, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)oxolan-2-yl]methyl (14R)-14-hydroxy-15,15-dimethyl-4,9,13-trioxo-1-(5-phenyl-1,2-oxazol-3-yl)-2-thia-5,8,12-triazahexadecan-16-yl dihydrogen diphosphate | | Authors: | Re, R.N, Noel, J.P, Burkart, M.D. | | Deposit date: | 2024-06-02 | | Release date: | 2025-05-14 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Elucidating the Iterative Elongation Mechanism in a Type III Polyketide Synthase.
J.Am.Chem.Soc., 147, 2025
|
|
8DLP
 
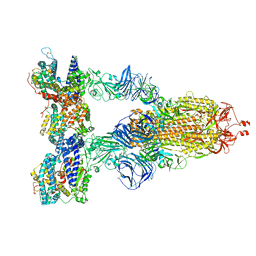 | | Cryo-EM structure of SARS-CoV-2 Gamma (P.1) spike protein in complex with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2022-08-31 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | SARS-CoV-2 variants of concern: spike protein mutational analysis and epitope for broad neutralization.
Nat Commun, 13, 2022
|
|
8DLX
 
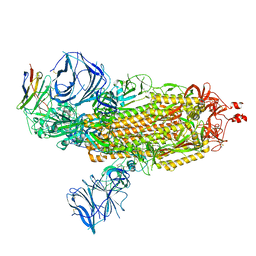 | | Cryo-EM structure of SARS-CoV-2 Epsilon (B.1.429) spike protein in complex with VH ab6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2022-08-31 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.45 Å) | | Cite: | SARS-CoV-2 variants of concern: spike protein mutational analysis and epitope for broad neutralization.
Nat Commun, 13, 2022
|
|
5S1C
 
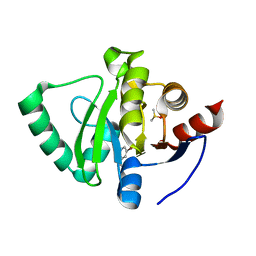 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with Z3034471507 | | Descriptor: | 1-(5-bromopyridin-3-yl)methanamine, DIMETHYL SULFOXIDE, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.174 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5XHW
 
 | | Crystal structure of HddC from Yersinia pseudotuberculosis | | Descriptor: | Putative 6-deoxy-D-mannoheptose pathway protein, SULFATE ION | | Authors: | Park, J, Kim, H, Kim, S, Shin, D.H. | | Deposit date: | 2017-04-24 | | Release date: | 2018-04-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of d-glycero-alpha-d-manno-heptose-1-phosphate guanylyltransferase from Yersinia pseudotuberculosis.
Biochim. Biophys. Acta, 1866, 2018
|
|
4XSL
 
 | | Crystal strcutre of D-tagatose 3-epimerase C66S from Pseudomonas cichorii in complex with glycerol | | Descriptor: | D-tagatose 3-epimerase, GLYCEROL, MANGANESE (II) ION | | Authors: | Yoshida, H, Yoshihara, A, Ishii, T, Izumori, K, Kamitori, S. | | Deposit date: | 2015-01-22 | | Release date: | 2016-01-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray structures of the Pseudomonas cichorii D-tagatose 3-epimerase mutant form C66S recognizing deoxy sugars as substrates
Appl. Microbiol. Biotechnol., 100, 2016
|
|
7K8G
 
 | | Crystal structure of bovine RPE65 in complex with 4-fluoro-MB-004 and palmitate | | Descriptor: | (1R)-3-amino-1-{4-fluoro-3-[(2-propylpentyl)oxy]phenyl}propan-1-ol, FE (II) ION, PALMITIC ACID, ... | | Authors: | Kiser, P.D. | | Deposit date: | 2020-09-27 | | Release date: | 2021-06-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Rational Alteration of Pharmacokinetics of Chiral Fluorinated and Deuterated Derivatives of Emixustat for Retinal Therapy.
J.Med.Chem., 64, 2021
|
|
5XGT
 
 | |
5BR7
 
 | | Structure of UDP-galactopyranose mutase from Corynebacterium diphtheriae in complex with citrate ion | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Wangkanont, K, Kiessling, L.L, Forest, K.T. | | Deposit date: | 2015-05-29 | | Release date: | 2016-06-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Conformational Control of UDP-Galactopyranose Mutase Inhibition.
Biochemistry, 56, 2017
|
|
7K89
 
 | |
3L7L
 
 | | Structure of the Wall Teichoic Acid Polymerase TagF, H444N + CDPG (30 minute soak) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, SULFATE ION, ... | | Authors: | Lovering, A.L, Strynadka, N.C.J. | | Deposit date: | 2009-12-28 | | Release date: | 2010-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure of the bacterial teichoic acid polymerase TagF provides insights into membrane association and catalysis.
Nat.Struct.Mol.Biol., 17, 2010
|
|
6D5W
 
 | | Ras:SOS:Ras in complex with a small molecule activator | | Descriptor: | 10-[(4-fluorophenyl)methyl]-2,3,4,10-tetrahydropyrimido[1,2-a]benzimidazole, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Phan, J, Hodges, T, Fesik, S.W. | | Deposit date: | 2018-04-19 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.478 Å) | | Cite: | Discovery and Structure-Based Optimization of Benzimidazole-Derived Activators of SOS1-Mediated Nucleotide Exchange on RAS.
J. Med. Chem., 61, 2018
|
|
