4RGW
 
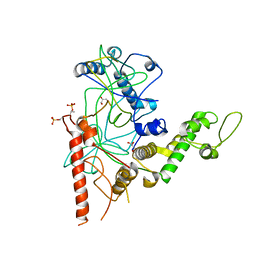 | | Crystal Structure of a TAF1-TAF7 Complex in Human Transcription Factor IID | | Descriptor: | GLYCEROL, Transcription initiation factor TFIID subunit 1, Transcription initiation factor TFIID subunit 7 | | Authors: | Wang, H, Curran, E.C, Hinds, T.R, Wang, E.H, Zheng, N. | | Deposit date: | 2014-09-30 | | Release date: | 2014-12-03 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Crystal structure of a TAF1-TAF7 complex in human transcription factor IID reveals a promoter binding module.
Cell Res., 24, 2014
|
|
6QMM
 
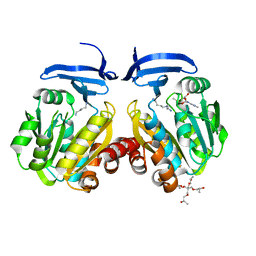 | | Crystal structure of Synecochoccus Spermidine Synthase in complex with putrescine, spermidine and MTA | | Descriptor: | (2S)-1-[3-{[(2R)-2-hydroxypropyl]oxy}-2,2-bis({[(2R)-2-hydroxypropyl]oxy}methyl)propoxy]propan-2-ol, 1,4-DIAMINOBUTANE, 5'-DEOXY-5'-METHYLTHIOADENOSINE, ... | | Authors: | Guedez, G, Pothipongsa, A, Incharoensakdi, A, Salminen, T.A. | | Deposit date: | 2019-02-07 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Crystal structure of dimericSynechococcusspermidine synthase with bound polyamine substrate and product.
Biochem.J., 476, 2019
|
|
7UKW
 
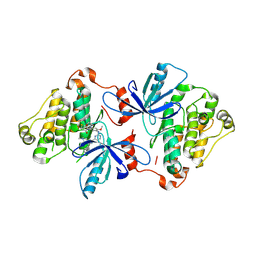 | | EGFR(T790M/V948R) in complex with Lazertinib (YH25448) | | Descriptor: | Epidermal growth factor receptor, N-[5-{[(4P)-4-{4-[(dimethylamino)methyl]-3-phenyl-1H-pyrazol-1-yl}pyrimidin-2-yl]amino}-4-methoxy-2-(morpholin-4-yl)phenyl]propanamide | | Authors: | Pham, C.D, Heppner, D.E. | | Deposit date: | 2022-04-02 | | Release date: | 2022-11-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for Inhibition of Mutant EGFR with Lazertinib (YH25448).
Acs Med.Chem.Lett., 13, 2022
|
|
5FO5
 
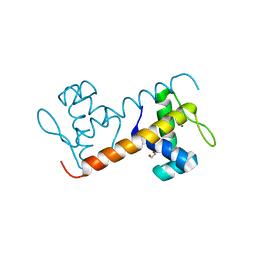 | | Structure of the DNA-binding domain of Escherichia coli methionine biosynthesis regulator MetR | | Descriptor: | 1,2-ETHANEDIOL, HTH-TYPE TRANSCRIPTIONAL REGULATOR METR, MAGNESIUM ION | | Authors: | Punekar, A.S, Porter, J, Urbanowski, M.L, Stauffer, G.V, Carr, S.B, Phillips, S.E. | | Deposit date: | 2015-11-18 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structural Basis for DNA Recognition by the Transcription Regulator Metr.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
5JN7
 
 | | Carbonic Anhydrase II In Complex WITH U-CH3 | | Descriptor: | 4-{[(3,5-dimethylphenyl)carbamoyl]amino}benzene-1-sulfonamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | McKenna, R, Mboge, M.Y, Mahon, B.P. | | Deposit date: | 2016-04-29 | | Release date: | 2017-05-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.517 Å) | | Cite: | Carbonic Anhydrase II In Complex WITH U-CH3
To Be Published
|
|
1A70
 
 | | SPINACH FERREDOXIN | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FERREDOXIN | | Authors: | Binda, C, Coda, A, Mattevi, A, Aliverti, A, Zanetti, G. | | Deposit date: | 1998-03-19 | | Release date: | 1998-11-25 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the mutant E92K of [2Fe-2S] ferredoxin I from Spinacia oleracea at 1.7 A resolution.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
8E4A
 
 | | Pseudomonas LpxC in complex with LPC-233 | | Descriptor: | 4-(4-cyclopropylbuta-1,3-diyn-1-yl)-N-[(2S,3S)-4,4-difluoro-3-hydroxy-1-(hydroxyamino)-3-methyl-1-oxobutan-2-yl]benzamide, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Najeeb, J, Zhou, P. | | Deposit date: | 2022-08-17 | | Release date: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.034 Å) | | Cite: | Preclinical safety and efficacy characterization of an LpxC inhibitor against Gram-negative pathogens.
Sci Transl Med, 15, 2023
|
|
4OAE
 
 | | Crystal structure of a GNAT superfamily acetyltransferase PA4794 C29A/C117A/Y128A mutant in complex with chloramphenicol | | Descriptor: | 1,2-ETHANEDIOL, CHLORAMPHENICOL, GNAT superfamily acetyltransferase PA4794, ... | | Authors: | Majorek, K.A, Chruszcz, M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-01-04 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal structure of a GNAT superfamily acetyltransferase PA4794 C29A/C117A/Y128A mutant in complex with chloramphenicol
To be Published
|
|
7YRE
 
 | | Crystal structure of a bright green fluorescent protein (StayGold) with triple mutations (N137A, Q140S, Y187F) in jellyfish Cytaeis uchidae from Biortus | | Descriptor: | 1,2-ETHANEDIOL, staygold(N137A,Q140S,Y187F) | | Authors: | Wu, J, Wang, F, Gui, W, Cheng, W, Yang, Y. | | Deposit date: | 2022-08-09 | | Release date: | 2023-08-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a bright green fluorescent protein (StayGold) with triple mutations (N137A, Q140S, Y187F) in jellyfish Cytaeis uchidae from Biortus
To Be Published
|
|
6TTE
 
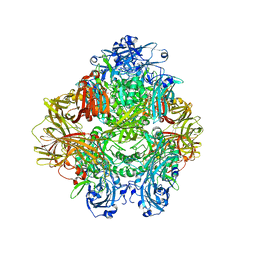 | | Beta-galactosidase in complex with PETG | | Descriptor: | 2-phenylethyl 1-thio-beta-D-galactopyranoside, Beta-galactosidase, MAGNESIUM ION | | Authors: | Saur, M, Hartshorn, M.J, Dong, J, Reeks, J, Bunkoczi, G, Jhoti, H, Williams, P.A. | | Deposit date: | 2019-12-27 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Fragment-based drug discovery using cryo-EM.
Drug Discov Today, 25, 2020
|
|
5DTF
 
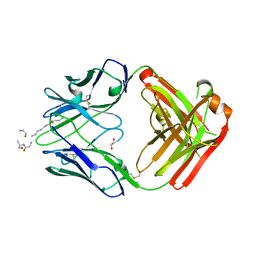 | |
9GE3
 
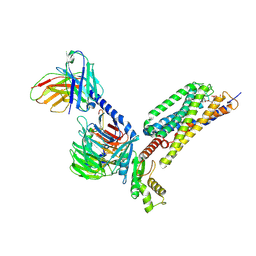 | | Structure of GPR55 in complex with G13 and endogenous lipid agonist lysophosphatidylinositol | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Green fluorescent protein,G-protein coupled receptor 55, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Claff, T, Ebenhoch, R, Weichert, D. | | Deposit date: | 2024-08-06 | | Release date: | 2025-03-05 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Structural basis for lipid-mediated activation of G protein-coupled receptor GPR55.
Nat Commun, 16, 2025
|
|
157L
 
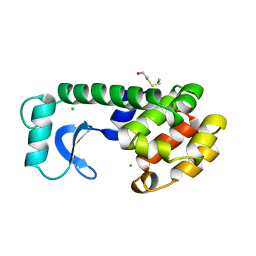 | | CONTROL OF ENZYME ACTIVITY BY AN ENGINEERED DISULFIDE BOND | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Blaber, M, Matthews, B.W. | | Deposit date: | 1994-06-20 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Alanine scanning mutagenesis of the alpha-helix 115-123 of phage T4 lysozyme: effects on structure, stability and the binding of solvent.
J.Mol.Biol., 246, 1995
|
|
8JJR
 
 | | Cryo-EM structure of Symbiodinium photosystem I | | Descriptor: | (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Zhao, L.S, Wang, N, Li, K, Zhang, Y.Z, Liu, L.N. | | Deposit date: | 2023-05-31 | | Release date: | 2024-05-15 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Architecture of symbiotic dinoflagellate photosystem I-light-harvesting supercomplex in Symbiodinium.
Nat Commun, 15, 2024
|
|
4ZL9
 
 | | Crystal structure of Pseudomonas aeruginosa DsbA E82I: Crystal III | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, TETRAETHYLENE GLYCOL, ... | | Authors: | McMahon, R.M, Martin, J.L. | | Deposit date: | 2015-05-01 | | Release date: | 2015-12-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Sent packing: protein engineering generates a new crystal form of Pseudomonas aeruginosa DsbA1 with increased catalytic surface accessibility.
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
6QXK
 
 | | Human PIM1 bound to OX0999 | | Descriptor: | 2-[(1-methylpiperidin-4-yl)methylamino]-5-[[2-[4-(trifluoromethyloxy)phenyl]-1,3-thiazol-4-yl]methyl]-1,3-thiazol-4-one, Pimtide, Serine/threonine-protein kinase pim-1 | | Authors: | Alexander, L.T, Elkins, J.M, Russell, A, Bountra, C, Edwards, A.M, Knapp, S. | | Deposit date: | 2019-03-07 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | PIM1 bound to OX0999
To Be Published
|
|
5FGU
 
 | | Structure of Sda1 nuclease apoprotein as an EGFP fixed-arm fusion | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Green fluorescent protein,Extracellular streptodornase D, ... | | Authors: | Moon, A.F, Krahn, J.M, Xun, L, Cuneo, M.J, Pedersen, L.C. | | Deposit date: | 2015-12-21 | | Release date: | 2016-03-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.896 Å) | | Cite: | Structural characterization of the virulence factor Sda1 nuclease from Streptococcus pyogenes.
Nucleic Acids Res., 44, 2016
|
|
8BGA
 
 | | Structure of Mpro in complex with FGA146 | | Descriptor: | 3C-like proteinase nsp5, 4-methoxy-~{N}-[(2~{S})-4-methyl-1-[[(2~{S})-4-nitro-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]butan-2-yl]amino]-1-oxidanylidene-pentan-2-yl]-1~{H}-indole-2-carboxamide | | Authors: | Medrano, F.J, Romero, A. | | Deposit date: | 2022-10-27 | | Release date: | 2023-11-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.982 Å) | | Cite: | Peptidyl nitroalkene inhibitors of main protease rationalized by computational and crystallographic investigations as antivirals against SARS-CoV-2.
Commun Chem, 7, 2024
|
|
7SOC
 
 | |
5FH0
 
 | | The structure of rat cytosolic PEPCK variant E89A complex with GTP | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GUANOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Johnson, T.A, Holyoak, T. | | Deposit date: | 2015-12-21 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Utilization of Substrate Intrinsic Binding Energy for Conformational Change and Catalytic Function in Phosphoenolpyruvate Carboxykinase.
Biochemistry, 55, 2016
|
|
8RUA
 
 | | Crystal structure of Rhizobium etli L-asparaginase ReAV C135A mutant | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Pokrywka, K, Grzechowiak, M, Sliwiak, J, Worsztynowicz, P, Loch, J.I, Ruszkowski, M, Gilski, M, Jaskolski, M. | | Deposit date: | 2024-01-30 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Probing the active site of Class 3 L-asparaginase by mutagenesis. I. Tinkering with the zinc coordination site of ReAV.
Front Chem, 12, 2024
|
|
8RUE
 
 | | Crystal structure of Rhizobium etli L-asparaginase ReAV H139A mutant | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Pokrywka, K, Grzechowiak, M, Sliwiak, J, Worsztynowicz, P, Loch, J.I, Ruszkowski, M, Gilski, M, Jaskolski, M. | | Deposit date: | 2024-01-30 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Probing the active site of Class 3 L-asparaginase by mutagenesis. I. Tinkering with the zinc coordination site of ReAV.
Front Chem, 12, 2024
|
|
6YBI
 
 | |
6YBO
 
 | |
6YBX
 
 | |
