1F1F
 
 | | CRYSTAL STRUCTURE OF CYTOCHROME C6 FROM ARTHROSPIRA MAXIMA | | Descriptor: | CYTOCHROME C6, HEME C | | Authors: | Kerfeld, C.A, Serag, A.A, Sawaya, M.R, Krogmann, D.W, Yeates, T.O. | | Deposit date: | 2000-05-18 | | Release date: | 2001-08-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of cytochrome c-549 and cytochrome c6 from the cyanobacterium Arthrospira maxima.
Biochemistry, 40, 2001
|
|
8GJV
 
 | |
1HYM
 
 | | HYDROLYZED TRYPSIN INHIBITOR (CMTI-V, MINIMIZED AVERAGE NMR STRUCTURE) | | Descriptor: | HYDROLYZED CUCURBITA MAXIMA TRYPSIN INHIBITOR V | | Authors: | Cai, M, Gong, Y, Prakash, O, Krishnamoorthi, R. | | Deposit date: | 1995-06-12 | | Release date: | 1995-09-15 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Reactive-site hydrolyzed Cucurbita maxima trypsin inhibitor-V: function, thermodynamic stability, and NMR solution structure.
Biochemistry, 34, 1995
|
|
1MIT
 
 | | RECOMBINANT CUCURBITA MAXIMA TRYPSIN INHIBITOR V (RCMTI-V) (NMR, MINIMIZED AVERAGE STRUCTURE) | | Descriptor: | TRYPSIN INHIBITOR V | | Authors: | Cai, M, Gong, Y, Huang, Y, Liu, J, Prakash, O, Wen, L, Wen, J.J, Huang, J.-K, Krishnamoorthi, R. | | Deposit date: | 1995-10-26 | | Release date: | 1996-04-03 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of recombinant Cucurbita maxima trypsin inhibitor-V determined by NMR spectroscopy.
Biochemistry, 35, 1996
|
|
1TIN
 
 | |
2I5M
 
 | |
2I5L
 
 | |
8OTT
 
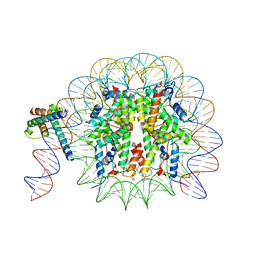 | | MYC-MAX bound to a nucleosome at SHL+5.8 | | Descriptor: | DNA (144-MER), Histone H2A type 1-B/E, Histone H2A type 1-K, ... | | Authors: | Stoos, L, Michael, A.K, Kempf, G, Kater, L, Cavadini, S, Thoma, N. | | Deposit date: | 2023-04-21 | | Release date: | 2023-05-24 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cooperation between bHLH transcription factors and histones for DNA access.
Nature, 619, 2023
|
|
2HAX
 
 | |
2ES2
 
 | |
7TC5
 
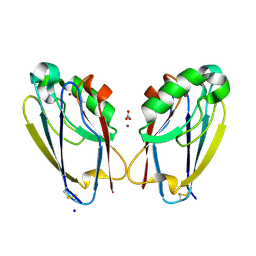 | | All Phe-Azurin variant - F15Y | | Descriptor: | Azurin, COPPER (II) ION, NITRATE ION, ... | | Authors: | Fedoretz-Maxwell, B.P, Worrall, L.J, Strynadka, N.C.J, Warren, J.J. | | Deposit date: | 2021-12-22 | | Release date: | 2022-06-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Impact of Second Coordination Sphere Methionine-Aromatic Interactions in Copper Proteins.
Inorg.Chem., 61, 2022
|
|
1FCG
 
 | | ECTODOMAIN OF HUMAN FC GAMMA RECEPTOR, FCGRIIA | | Descriptor: | PROTEIN (FC RECEPTOR FC(GAMMA)RIIA) | | Authors: | Maxwell, K.F, Powell, M.S, Garrett, T.P, Hogarth, P.M. | | Deposit date: | 1999-04-07 | | Release date: | 2000-04-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the human leukocyte Fc receptor, Fc gammaRIIa.
Nat.Struct.Biol., 6, 1999
|
|
8FZM
 
 | |
8GAI
 
 | |
7TC6
 
 | | All Phe-Azurin variant - F15W | | Descriptor: | Azurin, COPPER (II) ION, NITRATE ION | | Authors: | Fedoretz-Maxwell, B.P, Worrall, L.J, Strynadka, N.C.J, Warren, J.J. | | Deposit date: | 2021-12-22 | | Release date: | 2022-06-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Impact of Second Coordination Sphere Methionine-Aromatic Interactions in Copper Proteins.
Inorg.Chem., 61, 2022
|
|
7N8J
 
 | |
1HYW
 
 | | SOLUTION STRUCTURE OF BACTERIOPHAGE LAMBDA GPW | | Descriptor: | HEAD-TO-TAIL JOINING PROTEIN W | | Authors: | Maxwell, K.L, Yee, A.A, Booth, V, Arrowsmith, C.H, Gold, M, Davidson, A.R. | | Deposit date: | 2001-01-22 | | Release date: | 2001-04-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The solution structure of bacteriophage lambda protein W, a small morphogenetic protein possessing a novel fold.
J.Mol.Biol., 308, 2001
|
|
1K0H
 
 | | Solution structure of bacteriophage lambda gpFII | | Descriptor: | gpFII | | Authors: | Maxwell, K.L, Yee, A.A, Arrowsmith, C.H, Gold, M, Davidson, A.R. | | Deposit date: | 2001-09-19 | | Release date: | 2002-07-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the bacteriophage lambda head-tail joining protein, gpFII.
J.Mol.Biol., 318, 2002
|
|
7RZ3
 
 | |
2LTF
 
 | |
2BCX
 
 | | Crystal structure of calmodulin in complex with a ryanodine receptor peptide | | Descriptor: | CALCIUM ION, Calmodulin, Ryanodine receptor 1 | | Authors: | Maximciuc, A.A, Shamoo, Y, MacKenzie, K.R. | | Deposit date: | 2005-10-19 | | Release date: | 2006-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Complex of calmodulin with a ryanodine receptor target reveals a novel, flexible binding mode.
Structure, 14, 2006
|
|
8JRD
 
 | | Chalcone synthase from Glycine max (L.) Merr (soybean) complexed with naringenin and coenzyme A | | Descriptor: | 1,2-ETHANEDIOL, COENZYME A, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Waki, T, Imaizumi, R, Nakata, S, Yanai, T, Takeshita, K, Sakai, N, Kataoka, K, Yamamoto, M, Nakayama, T, Yamashita, S. | | Deposit date: | 2023-06-16 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural insights into catalytic promiscuity of chalcone synthase from Glycine max (L.) Merr.: Coenzyme A-induced alteration of product specificity.
Biochem.Biophys.Res.Commun., 718, 2024
|
|
8OTS
 
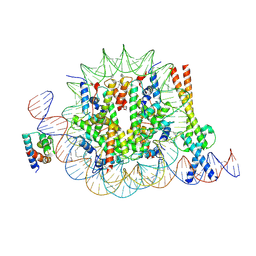 | | OCT4 and MYC-MAX co-bound to a nucleosome | | Descriptor: | DNA (127-MER), Green fluorescent protein,POU domain, class 5, ... | | Authors: | Michael, A.K, Stoos, L, Kempf, G, Cavadini, S, Thoma, N. | | Deposit date: | 2023-04-21 | | Release date: | 2023-05-24 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cooperation between bHLH transcription factors and histones for DNA access.
Nature, 619, 2023
|
|
8C8A
 
 | | The NMR structure of the MAX28 effector from Magnaporthe oryzae | | Descriptor: | But2 domain-containing protein | | Authors: | Lahfa, M, Padilla, A, de Guillen, K, Kroj, T, Roumestand, C, Barthe, P. | | Deposit date: | 2023-01-19 | | Release date: | 2024-02-14 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | The structural landscape and diversity of Pyricularia oryzae MAX effectors revisited.
Plos Pathog., 20, 2024
|
|
1YBA
 
 | | The active form of phosphoglycerate dehydrogenase | | Descriptor: | 2-OXOGLUTARIC ACID, D-3-phosphoglycerate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Thompson, J.R, Banaszak, L.J. | | Deposit date: | 2004-12-20 | | Release date: | 2005-04-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Vmax Regulation through Domain and Subunit Changes. The Active Form of Phosphoglycerate Dehydrogenase
Biochemistry, 44, 2005
|
|
