3SUI
 
 | |
6YUA
 
 | |
6YTT
 
 | | CO-dehydrogenase/Acetyl-CoA synthase (CODH/ACS) from Clostridium autoethanogenum at 3.0-A resolution | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, CO dehydrogenase/acetyl-CoA synthase complex, beta subunit, ... | | Authors: | Wagner, T, Lemaire, O.N. | | Deposit date: | 2020-04-24 | | Release date: | 2020-11-04 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Gas channel rerouting in a primordial enzyme: Structural insights of the carbon-monoxide dehydrogenase/acetyl-CoA synthase complex from the acetogen Clostridium autoethanogenum.
Biochim Biophys Acta Bioenerg, 1862, 2020
|
|
4G28
 
 | | Calcium-calmodulin complexed with the calmodulin binding domain from a small conductance potassium channel splice variant and EBIO-1 | | Descriptor: | 1-ethyl-1,3-dihydro-2H-benzimidazol-2-one, CALCIUM ION, Calmodulin, ... | | Authors: | Zhang, M, Pascal, J.M, Zhang, J.-F. | | Deposit date: | 2012-07-11 | | Release date: | 2012-09-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Identification of the functional binding pocket for compounds targeting small-conductance Ca(2+)-activated potassium channels.
Nat Commun, 3, 2012
|
|
5V02
 
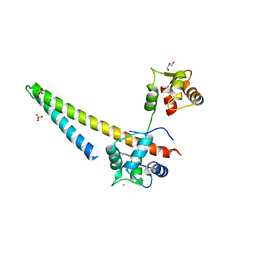 | |
1RJT
 
 | |
7AD3
 
 | | Class D GPCR Ste2 dimer coupled to two G proteins | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-factor mating pheromone, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Velazhahan, V, Tate, C. | | Deposit date: | 2020-09-14 | | Release date: | 2020-12-09 | | Last modified: | 2021-01-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the class D GPCR Ste2 dimer coupled to two G proteins.
Nature, 589, 2021
|
|
5V03
 
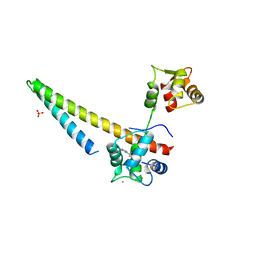 | |
5MX5
 
 | | Mouse PA28alpha-beta | | Descriptor: | PHOSPHATE ION, Proteasome activator complex subunit 1, Proteasome activator complex subunit 2 | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2017-01-20 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Mammalian Proteasome Activator PA28 Forms an Asymmetric alpha 4 beta 3 Complex.
Structure, 25, 2017
|
|
6O84
 
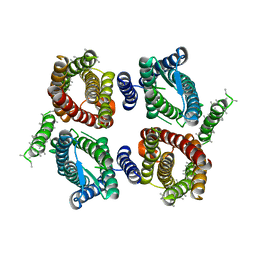 | | Cryo-EM structure of OTOP3 from xenopus tropicalis | | Descriptor: | LOC100127796 protein,LOC100127796 protein,OTOP3,LOC100127796 protein,LOC100127796 protein | | Authors: | Chen, Q.F, Bai, X.C, Jiang, Y.X. | | Deposit date: | 2019-03-08 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Structural and functional characterization of an otopetrin family proton channel.
Elife, 8, 2019
|
|
5WBX
 
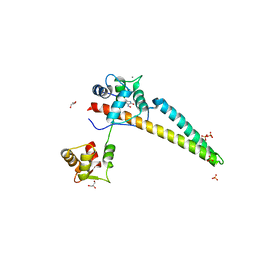 | |
5WC5
 
 | |
3HZZ
 
 | |
6ZHY
 
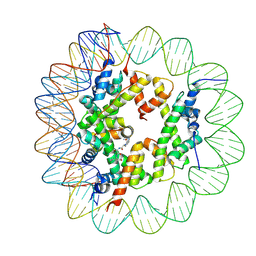 | | Cryo-EM structure of the regulatory linker of ALC1 bound to the nucleosome's acidic patch: hexasome class. | | Descriptor: | Chromodomain-helicase-DNA-binding protein 1-like, DNA (110-MER) Widom 601 sequence, Histone H2A type 1, ... | | Authors: | Bacic, L, Gaullier, G, Deindl, S. | | Deposit date: | 2020-06-24 | | Release date: | 2020-12-23 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mechanistic Insights into Regulation of the ALC1 Remodeler by the Nucleosome Acidic Patch.
Cell Rep, 33, 2020
|
|
2I5B
 
 | | The crystal structure of an ADP complex of Bacillus subtilis pyridoxal kinase provides evidence for the parralel emergence of enzyme activity during evolution | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Phosphomethylpyrimidine kinase | | Authors: | Newman, J.A, Das, S.K, Sedelnikova, S.E, Rice, D.W. | | Deposit date: | 2006-08-24 | | Release date: | 2006-09-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Crystal Structure of an ADP Complex of Bacillus subtilis Pyridoxal Kinase Provides Evidence for the Parallel Emergence of Enzyme Activity During Evolution.
J.Mol.Biol., 363, 2006
|
|
5MSK
 
 | | Mouse PA28beta | | Descriptor: | Proteasome activator complex subunit 2 | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2017-01-05 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The Mammalian Proteasome Activator PA28 Forms an Asymmetric alpha 4 beta 3 Complex.
Structure, 25, 2017
|
|
5VMS
 
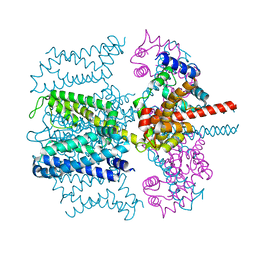 | | CryoEM structure of Xenopus KCNQ1 channel | | Descriptor: | CALCIUM ION, Calmodulin, Potassium voltage-gated channel subfamily KQT member 1 | | Authors: | Mackinnon, R, Sun, J. | | Deposit date: | 2017-04-28 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM Structure of a KCNQ1/CaM Complex Reveals Insights into Congenital Long QT Syndrome.
Cell, 169, 2017
|
|
6ZHX
 
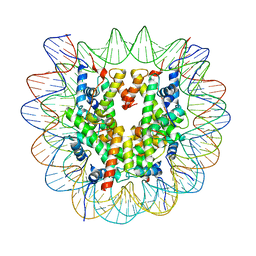 | | Cryo-EM structure of the regulatory linker of ALC1 bound to the nucleosome's acidic patch: nucleosome class. | | Descriptor: | Chromodomain-helicase-DNA-binding protein 1-like, DNA (145-MER) Widom 601 sequence, Histone H2A type 1, ... | | Authors: | Bacic, L, Gaullier, G, Croll, T.I, Deindl, S. | | Deposit date: | 2020-06-24 | | Release date: | 2020-12-23 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Mechanistic Insights into Regulation of the ALC1 Remodeler by the Nucleosome Acidic Patch.
Cell Rep, 33, 2020
|
|
7AJK
 
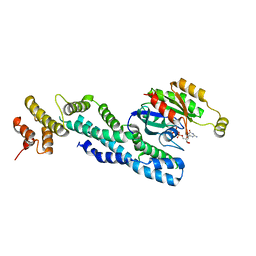 | | Crystal structure of CRYI-B Rac1 complex | | Descriptor: | CYFIP-related Rac1 interactor B, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Yelland, T, Anh, L, Insall, R, Machesky, L, Ismail, S. | | Deposit date: | 2020-09-29 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis of CYRI-B Direct Competition with Scar/WAVE Complex for Rac1.
Structure, 29, 2021
|
|
2BE4
 
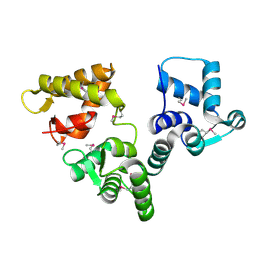 | | X-RAY STRUCTURE AN EF-HAND PROTEIN FROM DANIO RERIO Dr.36843 | | Descriptor: | hypothetical protein LOC449832 | | Authors: | Wesenberg, G.E, Phillips Jr, G.N, Han, B.W, Bitto, E, Bingman, C.A, Bae, E, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-10-21 | | Release date: | 2005-11-01 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray structure of Danio rerio secretagogin: A hexa-EF-hand calcium sensor.
Proteins, 76, 2009
|
|
9F0E
 
 | |
5NIN
 
 | |
4G27
 
 | | Calcium-calmodulin complexed with the calmodulin binding domain from a small conductance potassium channel splice variant and phenylurea | | Descriptor: | 1-phenylurea, CALCIUM ION, Calmodulin, ... | | Authors: | Zhang, M, Pascal, J.M, Zhang, J.-F. | | Deposit date: | 2012-07-11 | | Release date: | 2012-09-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Identification of the functional binding pocket for compounds targeting small-conductance Ca(2+)-activated potassium channels.
Nat Commun, 3, 2012
|
|
2ERJ
 
 | | Crystal structure of the heterotrimeric interleukin-2 receptor in complex with interleukin-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Debler, E.W, Stauber, D.J, Wilson, I.A. | | Deposit date: | 2005-10-25 | | Release date: | 2006-02-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the IL-2 signaling complex: Paradigm for a heterotrimeric cytokine receptor.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
7OIC
 
 | | Cryo-EM structure of late human 39S mitoribosome assembly intermediates, state 4 | | Descriptor: | 16S rRNA, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Cheng, J, Berninghausen, O, Beckmann, R. | | Deposit date: | 2021-05-11 | | Release date: | 2021-09-15 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A distinct assembly pathway of the human 39S late pre-mitoribosome.
Nat Commun, 12, 2021
|
|
