2QFK
 
 | | X-ray Crystal Structure Analysis of the Binding Site in the Ferric and Oxyferrous Forms of the Recombinant Heme Dehaloperoxidase Cloned from Amphitrite ornata | | Descriptor: | AMMONIUM ION, Dehaloperoxidase A, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | de Serrano, V.S, Chen, Z, Davis, M.F, Franzen, S. | | Deposit date: | 2007-06-27 | | Release date: | 2008-07-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | X-ray crystal structural analysis of the binding site in the ferric and
oxyferrous forms of the recombinant heme dehaloperoxidase cloned from Amphitrite ornata
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2QFL
 
 | | Structure of SuhB: Inositol monophosphatase and extragenic suppressor from E. coli | | Descriptor: | ACETATE ION, ETHYL ACETATE, Inositol-1-monophosphatase | | Authors: | Wang, Y, Stieglitz, K.A, Bubunenko, M, Court, D, Stec, B, Roberts, M.F. | | Deposit date: | 2007-06-27 | | Release date: | 2007-07-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of the R184A mutant of the inositol monophosphatase encoded by suhB and implications for its functional interactions in Escherichia coli.
J.Biol.Chem., 282, 2007
|
|
2QFN
 
 | | X-ray Crystal Structure Analysis of the Binding Site in the Ferric and Oxyferrous Forms of the Recombinant Heme Dehaloperoxidase Cloned from Amphitrite ornata | | Descriptor: | AMMONIUM ION, Dehaloperoxidase A, OXYGEN MOLECULE, ... | | Authors: | de Serrano, V, Chen, Z, Davis, M.F, Franzen, S. | | Deposit date: | 2007-06-27 | | Release date: | 2008-05-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | X-ray crystal structural analysis of the binding site in the ferric and oxyferrous forms of the recombinant heme dehaloperoxidase cloned from Amphitrite ornata
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2QFO
 
 | | HSP90 complexed with A143571 and A516383 | | Descriptor: | (3E)-3-[(phenylamino)methylidene]dihydrofuran-2(3H)-one, 4-METHYL-6-(TRIFLUOROMETHYL)PYRIMIDIN-2-AMINE, Heat shock protein HSP 90-alpha | | Authors: | Park, C.H. | | Deposit date: | 2007-06-27 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Discovery and design of novel HSP90 inhibitors using multiple fragment-based design strategies.
CHEM.BIOL.DRUG DES., 70, 2007
|
|
2QFP
 
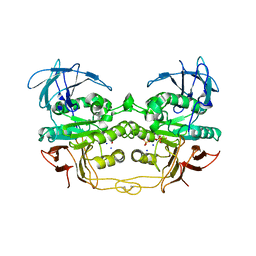 | | Crystal structure of red kidney bean purple acid phosphatase in complex with fluoride | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FE (III) ION, FLUORIDE ION, ... | | Authors: | Guddat, L.W, Schenk, G.S, Gahan, L.R, Elliot, T.W, Leung, E. | | Deposit date: | 2007-06-27 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of a purple acid phosphatase, representing different steps of this enzyme's catalytic cycle.
Bmc Struct.Biol., 8, 2008
|
|
2QFQ
 
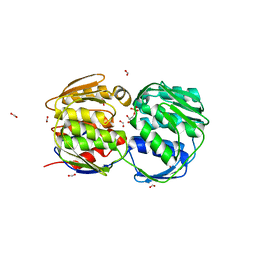 | | E. coli EPSP synthase Pro101Leu liganded with S3P | | Descriptor: | 3-phosphoshikimate 1-carboxyvinyltransferase, FORMIC ACID, SHIKIMATE-3-PHOSPHATE | | Authors: | Schonbrunn, E, Healy-Fried, M.L. | | Deposit date: | 2007-06-27 | | Release date: | 2007-10-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of glyphosate tolerance resulting from mutations of Pro101 in Escherichia coli 5-enolpyruvylshikimate-3-phosphate synthase.
J.Biol.Chem., 282, 2007
|
|
2QFR
 
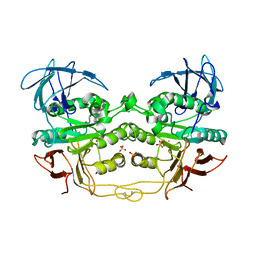 | | Crystal structure of red kidney bean purple acid phosphatase with bound sulfate | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, FE (III) ION, ... | | Authors: | Guddat, L.W, Schenk, G, Gahan, L.R, Elliot, T.W, Leung, E. | | Deposit date: | 2007-06-27 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of a purple acid phosphatase, representing different steps of this enzyme's catalytic cycle.
Bmc Struct.Biol., 8, 2008
|
|
2QFS
 
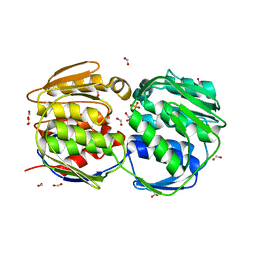 | | E.coli EPSP synthase Pro101Ser liganded with S3P | | Descriptor: | 3-phosphoshikimate 1-carboxyvinyltransferase, FORMIC ACID, SHIKIMATE-3-PHOSPHATE | | Authors: | Schonbrunn, E, Healy-Fried, M.L. | | Deposit date: | 2007-06-28 | | Release date: | 2007-10-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural basis of glyphosate tolerance resulting from mutations of Pro101 in Escherichia coli 5-enolpyruvylshikimate-3-phosphate synthase.
J.Biol.Chem., 282, 2007
|
|
2QFT
 
 | |
2QFU
 
 | |
2QFV
 
 | |
2QFW
 
 | |
2QFX
 
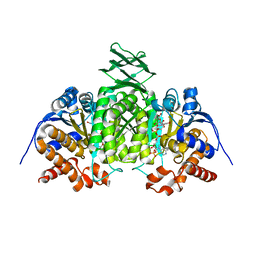 | |
2QFY
 
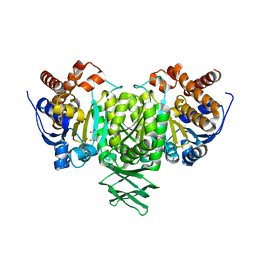 | |
2QFZ
 
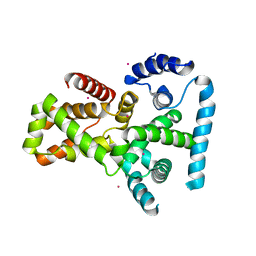 | | Crystal structure of human TBC1 domain family member 22A | | Descriptor: | TBC1 domain family member 22A, UNKNOWN ATOM OR ION | | Authors: | Tong, Y, Tempel, W, Dimov, S, Dong, A, Landry, R, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-06-28 | | Release date: | 2007-07-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of human TBC1 domain family member 22A.
To be Published
|
|
2QG0
 
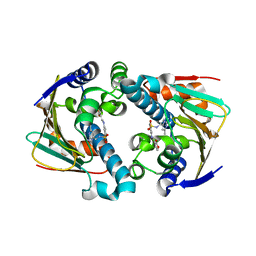 | | HSP90 complexed with A943037 | | Descriptor: | Heat shock protein HSP 90-alpha, N-[(2-AMINO-6-METHYLPYRIMIDIN-4-YL)METHYL]-3-{[(E)-(2-OXODIHYDROFURAN-3(2H)-YLIDENE)METHYL]AMINO}BENZENESULFONAMIDE | | Authors: | Park, C.H. | | Deposit date: | 2007-06-28 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery and design of novel HSP90 inhibitors using multiple fragment-based design strategies.
Chem.Biol.Drug Des., 70, 2007
|
|
2QG1
 
 | | Crystal structure of the 11th PDZ domain of MPDZ (MUPP1) | | Descriptor: | 1,2-ETHANEDIOL, Multiple PDZ domain protein | | Authors: | Papagrigoriou, E, Salah, E, Phillips, C, Savitsky, P, Boisguerin, P, Oschkinat, H, Gileadi, C, Yang, X, Elkins, J.M, Ugochukwu, E, Bunkoczi, G, Uppenberg, J, Sundstrom, M, Arrowsmith, C.H, Weigelt, J, Edwards, A, von Delft, F, Doyle, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-06-28 | | Release date: | 2007-07-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of the 11th PDZ domain of MPDZ (MUPP1).
To be Published
|
|
2QG2
 
 | | HSP90 complexed with A917985 | | Descriptor: | 3-({2-[(2-AMINO-6-METHYLPYRIMIDIN-4-YL)ETHYNYL]BENZYL}AMINO)-1,3-OXAZOL-2(3H)-ONE, Heat shock protein HSP 90-alpha | | Authors: | Park, C.H. | | Deposit date: | 2007-06-28 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery and design of novel HSP90 inhibitors using multiple fragment-based design strategies.
Chem.Biol.Drug Des., 70, 2007
|
|
2QG3
 
 | |
2QG4
 
 | | Crystal structure of human UDP-glucose dehydrogenase product complex with UDP-glucuronate | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Kavanagh, K.L, Guo, K, Bunkoczi, G, Savitsky, P, Pilka, E, Bhatia, C, Niesen, F, Smee, C, Berridge, G, von Delft, F, Weigelt, J, Arrowsmith, C.H, Sundstrom, M, Edwards, A, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-06-28 | | Release date: | 2007-07-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and mechanism of human UDP-glucose 6-dehydrogenase.
J.Biol.Chem., 286, 2011
|
|
2QG5
 
 | | Cryptosporidium parvum calcium dependent protein kinase cgd7_1840 | | Descriptor: | Calcium/calmodulin-dependent protein kinase | | Authors: | Lunin, V.V, Wernimont, A.K, Lew, J, Wasney, G, Kozieradzki, I, Vedadi, M, Bochkarev, A, Arrowsmith, C.H, Sundstrom, M, Weigelt, J, Edwards, A.E, Hui, R, Artz, J, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-06-28 | | Release date: | 2007-09-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cryptosporidium parvum calcium dependent protein kinase cgd7_1840
To be Published
|
|
2QG6
 
 | |
2QG7
 
 | | Plasmodium vivax ethanolamine kinase Pv091845 | | Descriptor: | SULFATE ION, ethanolamine kinase Pv091845 | | Authors: | Lunin, V.V, Wernimont, A.K, Mulichak, A, Lew, J, Wasney, G, Senisterra, G, Kozieradzki, I, Vedadi, M, Bochkarev, A, Arrowsmith, C.H, Sundstrom, M, Weigelt, J, Edwards, A.E, Hui, R, Hills, T, Artz, J, Xiao, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-06-28 | | Release date: | 2007-07-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.407 Å) | | Cite: | Plasmodium vivax ethanolamine kinase Pv091845
TO BE PUBLISHED
|
|
2QG8
 
 | | Plasmodium yoelii acyl carrier protein synthase PY06285 with ADP bound | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, Acyl Carrier Protein Synthase PY06285, MAGNESIUM ION | | Authors: | Lunin, V.V, Wernimont, A.K, Lew, J, Wasney, G, Kozieradzki, I, Vedadi, M, Bochkarev, A, Arrowsmith, C.H, Sundstrom, M, Weigelt, J, Edwards, A.E, Hui, R, Brokx, S, Altamentova, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-06-28 | | Release date: | 2007-07-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Plasmodium yoelii acyl carrier protein synthase PY06285 with ADP bound
TO BE PUBLISHED
|
|
2QG9
 
 | | Structure of a regulatory subunit mutant D19A of ATCase from E. coli | | Descriptor: | Aspartate carbamoyltransferase catalytic chain, Aspartate carbamoyltransferase regulatory chain, ZINC ION | | Authors: | Stec, B, Williams, M.K, Stieglitz, K.A, Kantrowitz, E.R. | | Deposit date: | 2007-06-28 | | Release date: | 2008-02-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Comparison of two T-state structures of regulatory-chain mutants of Escherichia coli aspartate transcarbamoylase suggests that His20 and Asp19 modulate the response to heterotropic effectors.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
