2V4B
 
 | | Crystal Structure of Human ADAMTS-1 catalytic Domain and Cysteine- Rich Domain (apo-form) | | Descriptor: | ADAMTS-1, CADMIUM ION, MAGNESIUM ION, ... | | Authors: | Gerhardt, S, Hassall, G, Hawtin, P, McCall, E, Flavell, L, Minshull, C, Hargreaves, D, Ting, A, Pauptit, R.A, Parker, A.E, Abbott, W.M. | | Deposit date: | 2007-06-28 | | Release date: | 2008-01-15 | | Last modified: | 2019-04-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Human Adamts-1 Reveal a Conserved Catalytic Domain and a Disintegrin-Like Domain with a Fold Homologous to Cysteine-Rich Domains.
J.Mol.Biol., 373, 2007
|
|
4GOH
 
 | |
4IP0
 
 | | X-Ray Structure of the Complex Uridine Phosphorylase from Vibrio cholerae with Phosphate Ion at 1.29 A Resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, ETHANOL, ... | | Authors: | Prokofev, I.I, Lashkov, A.A, Gabdoulkhakov, A.G, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2013-01-09 | | Release date: | 2014-08-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.294 Å) | | Cite: | X-ray structures of uridine phosphorylase from Vibrio cholerae in complexes with uridine, thymidine, uracil, thymine, and phosphate anion: Substrate specificity of bacterial uridine phosphorylases
Crystallography Reports, 61, 2016
|
|
2WY0
 
 | | Crystal structure of mouse angiotensinogen in the oxidised form with space group P6122 | | Descriptor: | 1,2-ETHANEDIOL, ANGIOTENSINOGEN, SODIUM ION | | Authors: | Zhou, A, Wei, Z, Carrell, R.W, Read, R.J. | | Deposit date: | 2009-11-11 | | Release date: | 2010-10-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | A Redox Switch in Angiotensinogen Modulates Angiotensin Release.
Nature, 468, 2010
|
|
2WNT
 
 | | Crystal Structure of the Human Ribosomal protein S6 kinase | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, RIBOSOMAL PROTEIN S6 KINASE, ... | | Authors: | Muniz, J.R.C, Elkins, J.M, Wang, J, Ugochukwu, E, Salah, E, King, O, Picaud, S, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Knapp, S. | | Deposit date: | 2009-07-20 | | Release date: | 2009-08-25 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the Human Ribosomal Protein S6 Kinase
To be Published
|
|
4IGU
 
 | |
1T4B
 
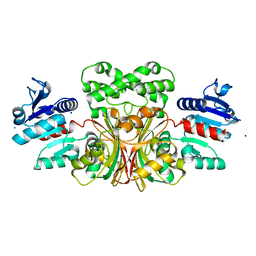 | | 1.6 Angstrom structure of Esherichia coli aspartate-semialdehyde dehydrogenase. | | Descriptor: | Aspartate-semialdehyde dehydrogenase, SODIUM ION | | Authors: | Nichols, C.E, Dhaliwal, B, Lockyer, M, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2004-04-29 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High-resolution Structures Reveal Details of Domain Closure and "Half-of-sites-reactivity" in Escherichia coli Aspartate beta-Semialdehyde Dehydrogenase.
J.Mol.Biol., 341, 2004
|
|
2UZZ
 
 | | X-ray structure of N-methyl-L-tryptophan oxidase (MTOX) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, N-METHYL-L-TRYPTOPHAN OXIDASE, SODIUM ION | | Authors: | Ilari, A, Fiorillo, A, Franceschini, S, Bonamore, A, Colotti, G, Boffi, A. | | Deposit date: | 2007-05-03 | | Release date: | 2008-01-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The X-Ray Structure of N-Methyltryptophan Oxidase Reveals the Structural Determinants of Substrate Specificity.
Proteins, 71, 2008
|
|
4GY7
 
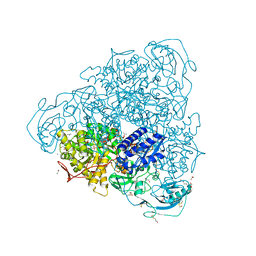 | | Crystallographic structure analysis of urease from Jack bean (Canavalia ensiformis) at 1.49 A Resolution | | Descriptor: | 1,2-ETHANEDIOL, ACETONE, BETA-MERCAPTOETHANOL, ... | | Authors: | Begum, A, Banumathi, S, Choudhary, M.I, Betzel, C. | | Deposit date: | 2012-09-05 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.492 Å) | | Cite: | Crystallographic structure analysis of urease from Jack bean (Canavalia ensiformis) at 1.49 A Resolution
To be Published
|
|
1A14
 
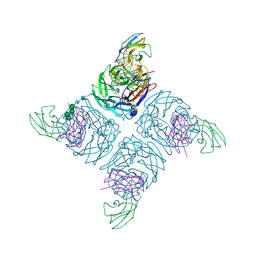 | | COMPLEX BETWEEN NC10 ANTI-INFLUENZA VIRUS NEURAMINIDASE SINGLE CHAIN ANTIBODY WITH A 5 RESIDUE LINKER AND INFLUENZA VIRUS NEURAMINIDASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, NC10 FV (HEAVY CHAIN), ... | | Authors: | Malby, R.L, Mccoy, A.J, Kortt, A.A, Hudson, P.J, Colman, P.M. | | Deposit date: | 1997-12-21 | | Release date: | 1998-05-13 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three-dimensional structures of single-chain Fv-neuraminidase complexes.
J.Mol.Biol., 279, 1998
|
|
4H41
 
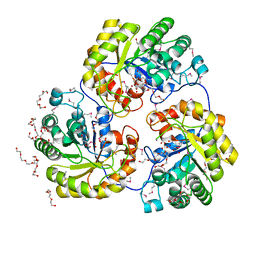 | |
4GNM
 
 | |
2WO0
 
 | | EDTA treated E. coli copper amine oxidase | | Descriptor: | COPPER (II) ION, PRIMARY AMINE OXIDASE, SODIUM ION | | Authors: | Smith, M.A, Pirrat, P, Pearson, A.R, Knowles, P.F, Phillips, S.E.V, McPherson, M.J. | | Deposit date: | 2009-07-21 | | Release date: | 2010-05-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Exploring the Roles of the Metal Ions in Escherichia Coli Copper Amine Oxidase.
Biochemistry, 49, 2010
|
|
2WOF
 
 | | EDTA treated E. coli copper amine oxidase | | Descriptor: | COPPER (II) ION, PRIMARY AMINE OXIDASE, SODIUM ION | | Authors: | Smith, M.A, Pirrat, P, Pearson, A.R, Knowles, P.F, Phillips, S.E.V, McPherson, M.J. | | Deposit date: | 2009-07-23 | | Release date: | 2010-05-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Exploring the Roles of the Metal Ions in Escherichia Coli Copper Amine Oxidase.
Biochemistry, 49, 2010
|
|
4GVO
 
 | | Putative L-Cystine ABC transporter from Listeria monocytogenes | | Descriptor: | CHLORIDE ION, HISTIDINE, Lmo2349 protein, ... | | Authors: | Osipiuk, J, Makowska-Grzyska, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-08-30 | | Release date: | 2012-09-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.448 Å) | | Cite: | Putative L-Cystine ABC transporter from Listeria monocytogenes
To be Published
|
|
3F5M
 
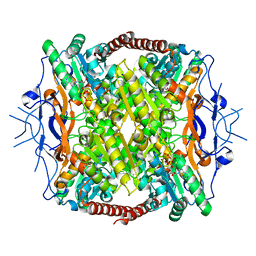 | | Crystal Structure of ATP-Bound Phosphofructokinase from Trypanosoma brucei | | Descriptor: | 6-phospho-1-fructokinase (ATP-dependent phosphofructokinase), ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, ... | | Authors: | McNae, I.W, Martinez-Oyanedel, J, Keillor, J.W, Michels, P.A.M, Fothergill-Gilmore, L.A, Walkinshaw, M.D. | | Deposit date: | 2008-11-04 | | Release date: | 2008-11-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of ATP-bound phosphofructokinase from Trypanosoma brucei reveals conformational transitions different from those of other phosphofructokinases.
J.Mol.Biol., 385, 2009
|
|
3FF1
 
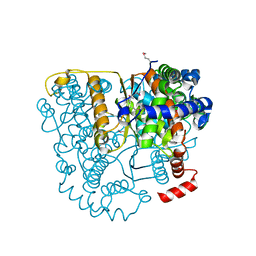 | | Structure of Glucose 6-phosphate Isomerase from Staphylococcus aureus | | Descriptor: | GLUCOSE-6-PHOSPHATE, Glucose-6-phosphate isomerase, SODIUM ION | | Authors: | Anderson, S.M, Brunzelle, J.S, Onopriyenko, O, Peterson, S, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-12-01 | | Release date: | 2009-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of Glucose 6-phosphate Isomerase from Staphylococcus aureus
TO BE PUBLISHED
|
|
3FW4
 
 | |
4I0W
 
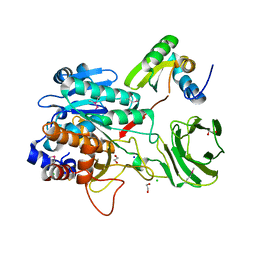 | | Structure of the Clostridium Perfringens CspB protease | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Protease CspB, ... | | Authors: | Adams, C.M, Eckenroth, B.E, Doublie, S. | | Deposit date: | 2012-11-19 | | Release date: | 2013-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and functional analysis of the CspB protease required for Clostridium spore germination.
Plos Pathog., 9, 2013
|
|
3G3F
 
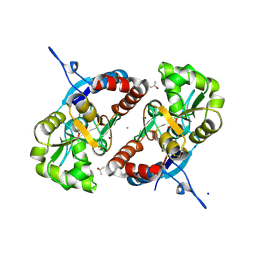 | |
4IA6
 
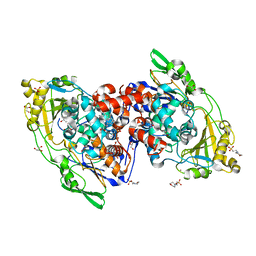 | | Hydratase from lactobacillus acidophilus in a ligand bound form (LA LAH) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Khoshnevis, S, Neumann, P, Ficner, R. | | Deposit date: | 2012-12-06 | | Release date: | 2013-03-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure analysis of a fatty acid double-bond hydratase from Lactobacillus acidophilus
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4I9B
 
 | | Structure of aminoaldehyde dehydrogenase 1 from Solanum lycopersium (SlAMADH1) with a thiohemiacetal intermediate | | Descriptor: | (2-hydroxyethoxy)acetaldehyde, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Morera, S, Vigouroux, A, Kopecny, D. | | Deposit date: | 2012-12-05 | | Release date: | 2013-02-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Plant ALDH10 family: identifying critical residues for substrate specificity and trapping a thiohemiacetal intermediate.
J.Biol.Chem., 288, 2013
|
|
4J02
 
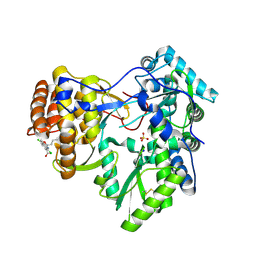 | | Crystal structure of hcv ns5b polymerase in complex with [(1R)-5,8-DICHLORO-1-PROPYL-1,3,4,9-TETRAHYDROPYRANO[3,4-B]INDOL-1-YL]ACETIC ACID | | Descriptor: | Genome polyprotein, SODIUM ION, SULFATE ION, ... | | Authors: | Coulombe, R. | | Deposit date: | 2013-01-30 | | Release date: | 2013-04-17 | | Last modified: | 2013-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of a novel series of non-nucleoside thumb pocket 2 HCV NS5B polymerase inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4J1H
 
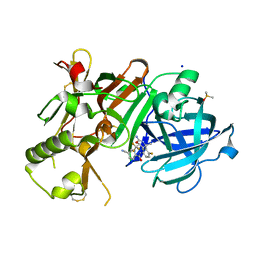 | | CRYSTAL STRUCTURE OF BACE-1 IN COMPLEX WITH 5-Cyano-pyridine-2-carboxylic acid [3-((4S,6R)-2-amino-4-methyl-6-trifluoromethyl-5,6-dihydro-4H-[1,3]oxazin-4-yl)-4-fluoro-phenyl]-amide | | Descriptor: | Beta-secretase 1, DIMETHYL SULFOXIDE, N-{3-[(4S,6R)-2-amino-4-methyl-6-(trifluoromethyl)-5,6-dihydro-4H-1,3-oxazin-4-yl]-4-fluorophenyl}-5-cyanopyridine-2-carboxamide, ... | | Authors: | Kuglstatter, A, Stihle, M. | | Deposit date: | 2013-02-01 | | Release date: | 2013-05-01 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | beta-Secretase (BACE1) Inhibitors with High In Vivo Efficacy Suitable for Clinical Evaluation in Alzheimer s Disease
J.Med.Chem., 56, 2013
|
|
4J1K
 
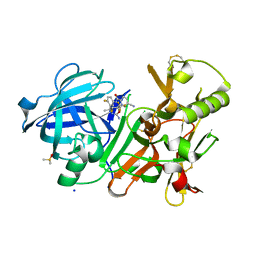 | | CRYSTAL STRUCTURE OF BACE-1 IN COMPLEX WITH 5-Cyano-pyridine-2-carboxylic acid [3-((4R,5R,6S)-2-amino-5-fluoro-4-methyl-6-trifluoromethyl-5,6-dihydro-4H-[1,3]oxazin-4-yl)-4-fluoro-phenyl]-amide | | Descriptor: | Beta-secretase 1, DIMETHYL SULFOXIDE, N-{3-[(4R,5R,6S)-2-amino-5-fluoro-4-methyl-6-(trifluoromethyl)-5,6-dihydro-4H-1,3-oxazin-4-yl]-4-fluorophenyl}-5-cyanopyridine-2-carboxamide, ... | | Authors: | Kuglstatter, A, Stihle, M. | | Deposit date: | 2013-02-01 | | Release date: | 2013-05-01 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | beta-Secretase (BACE1) Inhibitors with High In Vivo Efficacy Suitable for Clinical Evaluation in Alzheimer s Disease
J.Med.Chem., 56, 2013
|
|
